Cryo-EM Analysis of a Tri-Heme Cytochrome-Associated RC-LH1 Complex from the Marine Photoheterotrophic Bacterium Dinoroseobacter Shibae.
Wang, W., Liu, Y., Gu, J., An, S., Ma, C., Gao, H., Jiao, N., Shen, J.R., Beatty, J.T., Koblizek, M., Zhang, X., Zheng, Q., Chen, J.H.(2025) Adv Sci (Weinh) 12: e2413456-e2413456
- PubMed: 40112203
- DOI: https://doi.org/10.1002/advs.202413456
- Primary Citation of Related Structures:
8YY9, 8YZ2, 9KM0 - PubMed Abstract:
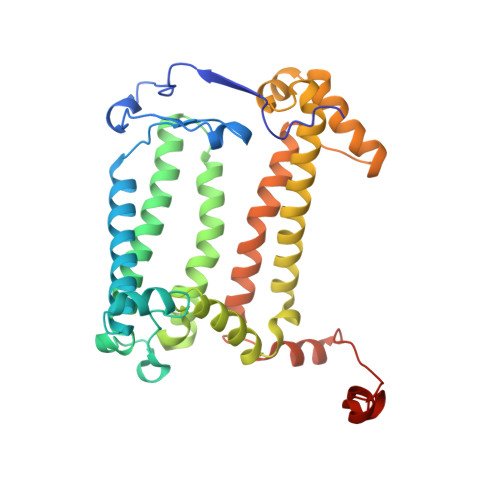
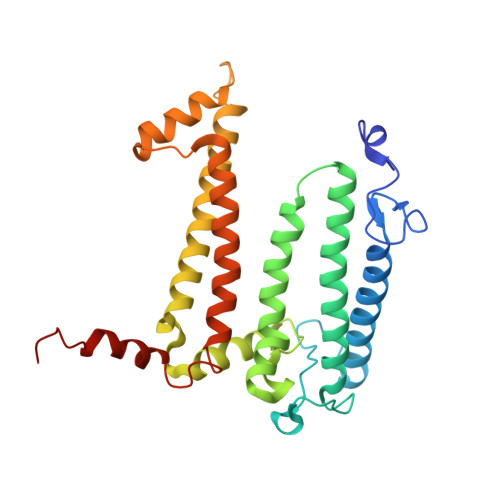
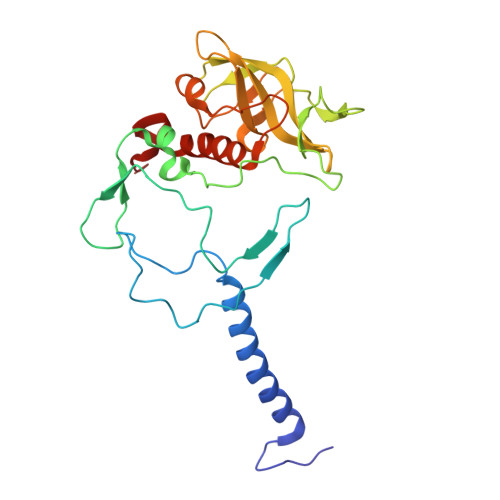
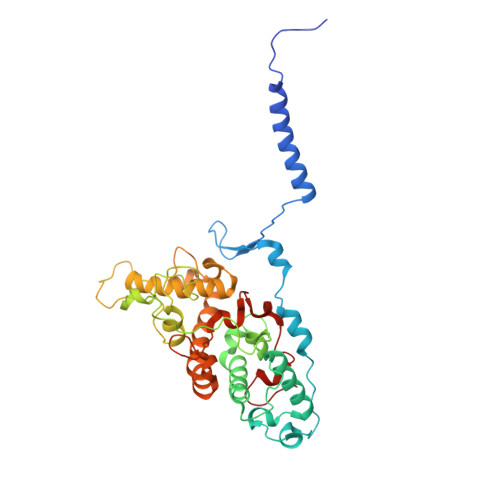
The reaction center-light harvesting 1 (RC-LH1) complex converts solar energy into electrical energy, driving the initiation of photosynthesis. The authors present a cryo-electron microscopy structure of the RC-LH1 isolated from a marine photoheterotrophic bacterium Dinoroseobacter shibae. The RC comprises four subunits, including a three-heme cytochrome (Cyt) c protein, and is surrounded by a closed LH ring composed of 17 pairs of antenna subunits. Notably, a novel subunit with an N-terminal "helix-turn-helix" motif embedded in the gap between the RC and the LH ring is identified. The purified RC-LH1 complex exhibits high stability in solutions containing Mg 2+ or Ca 2+ . The periplasmic Cyt c 2 is predicted to bind at the junction between the Cyt subunit and the membrane plane, enabling electron transfer from Cyt c 2 to the proximal heme of the tri-heme Cyt, and subsequently to the special pair of bacteriochlorophylls. These findings provide structural insights into the efficient energy and electron transfer processes within a distinct type of RC-LH1, and shed light on evolutionary adaptations of photosynthesis.
- College of Life Sciences, Zhejiang University, Hangzhou, Zhejiang, 310058, China.
Organizational Affiliation: