Cryo-EM structure of OXGR1 bound to leukotriene E4 and Gq proteins
Liu, A., Liu, Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| scFV16 | 247 | Vicugna pacos | Mutation(s): 0 | 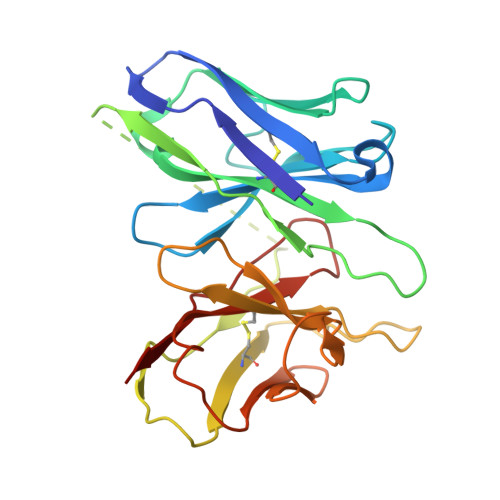 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | 339 | Homo sapiens | Mutation(s): 0 Gene Names: GNB1 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P62873 (Homo sapiens) Explore P62873 Go to UniProtKB: P62873 | |||||
PHAROS: P62873 GTEx: ENSG00000078369 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62873 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine nucleotide-binding protein G(q) subunit alpha | 354 | Homo sapiens | Mutation(s): 2 Gene Names: GNAI1, GNAQ, GAQ EC: 3.6.5 | 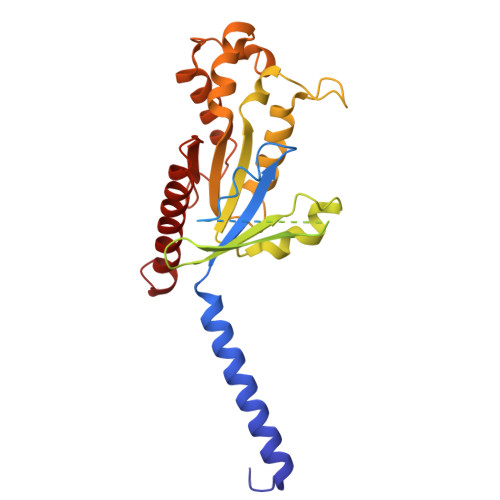 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P63096 (Homo sapiens) Explore P63096 Go to UniProtKB: P63096 | |||||
PHAROS: P63096 GTEx: ENSG00000127955 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P63096 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 | 71 | Homo sapiens | Mutation(s): 0 Gene Names: GNG2 | 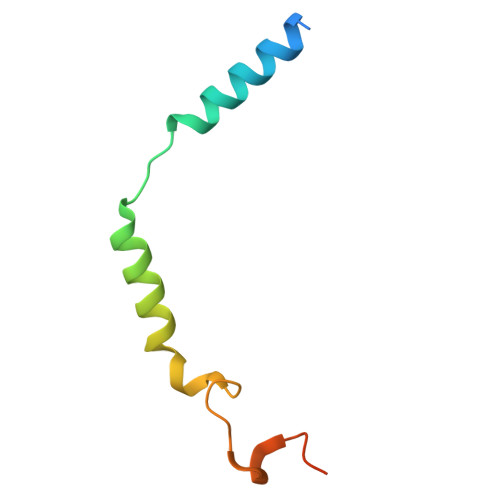 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P59768 (Homo sapiens) Explore P59768 Go to UniProtKB: P59768 | |||||
PHAROS: P59768 GTEx: ENSG00000186469 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P59768 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 2-oxoglutarate receptor 1 | 337 | Homo sapiens | Mutation(s): 0 Gene Names: OXGR1, GPR80, GPR99, P2RY15, P2Y15 | 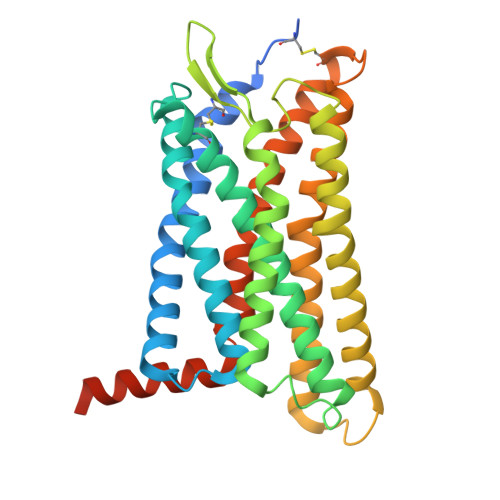 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q96P68 (Homo sapiens) Explore Q96P68 Go to UniProtKB: Q96P68 | |||||
PHAROS: Q96P68 GTEx: ENSG00000165621 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96P68 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| A1D7P (Subject of Investigation/LOI) Query on A1D7P | G [auth E] | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-azanyl-3-oxidanyl-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid C23 H37 N O5 S OTZRAYGBFWZKMX-FRFVZSDQSA-N |  | ||
| CLR Query on CLR | F [auth E] | CHOLESTEROL C27 H46 O HVYWMOMLDIMFJA-DPAQBDIFSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |