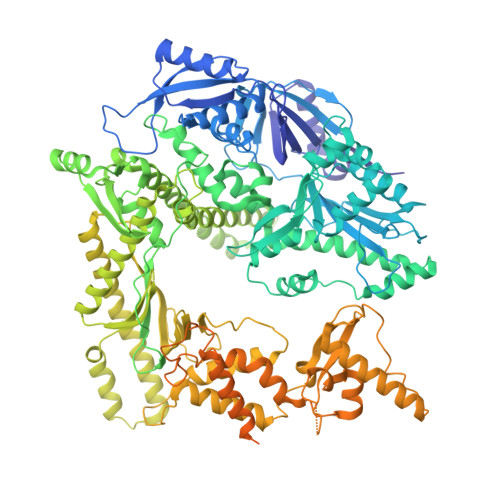
Cryo-EM structure of DNA polymerase of African swine fever virus.
Kuai, L., Sun, J., Peng, Q., Zhao, X., Yuan, B., Liu, S., Bi, Y., Shi, Y.(2024) Nucleic Acids Res 52: 10717-10729
- PubMed: 39189451
- DOI: https://doi.org/10.1093/nar/gkae739
- Primary Citation of Related Structures:
8YWG, 8YWI, 8YWM - PubMed Abstract:
African swine fever virus (ASFV) is one of the most important causative agents of animal diseases and can cause highly fatal diseases in swine. ASFV DNA polymerase (DNAPol) is responsible for genome replication and highly conserved in all viral genotypes showing an ideal target for drug development. Here, we systematically determined the structures of ASFV DNAPol in apo, replicating and editing states. Structural analysis revealed that ASFV DNAPol had a classical right-handed structure and showed the highest similarity to the structure of human polymerase delta. Intriguingly, ASFV DNAPol has a much longer fingers subdomain, and the thumb and palm subdomain form a unique interaction that has never been seen. Mutagenesis work revealed that the loss of this unique interaction decreased the enzymatic activity. We also found that the β-hairpin of ASFV DNAPol is located below the template strand in the editing state, which is different from the editing structures of other known B family DNAPols with the β-hairpin above the template strand. It suggests that B family DNAPols have evolved two ways to facilitate the dsDNA unwinding during the transition from replicating into editing state. These findings figured out the working mechanism of ASFV DNAPol and will provide a critical structural basis for the development of antiviral drugs.
- CAS Key Laboratory of Pathogen Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: