Structure of Erabutoxin bound to Torpedo acetylcholine receptor gives insight to intramolecular selectivity
Chan, C.Y.L., Li, H.H., Hibbs, R.E., Kini, R.M.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Acetylcholine receptor subunit alpha | 437 | Tetronarce californica | Mutation(s): 0 Membrane Entity: Yes | 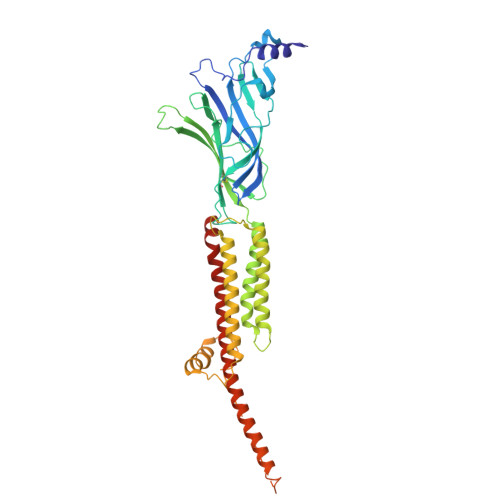 | |
UniProt | |||||
Find proteins for P02710 (Tetronarce californica) Explore P02710 Go to UniProtKB: P02710 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P02710 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Acetylcholine receptor subunit delta | 501 | Tetronarce californica | Mutation(s): 0 Membrane Entity: Yes | 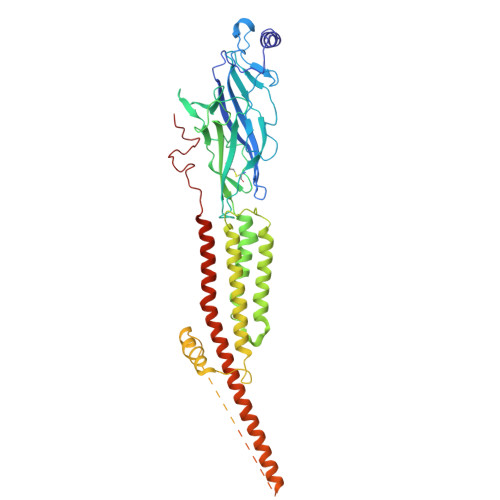 | |
UniProt | |||||
Find proteins for P02718 (Tetronarce californica) Explore P02718 Go to UniProtKB: P02718 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P02718 | ||||
Glycosylation | |||||
| Glycosylation Sites: 3 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Acetylcholine receptor subunit beta | 469 | Tetronarce californica | Mutation(s): 0 Membrane Entity: Yes | 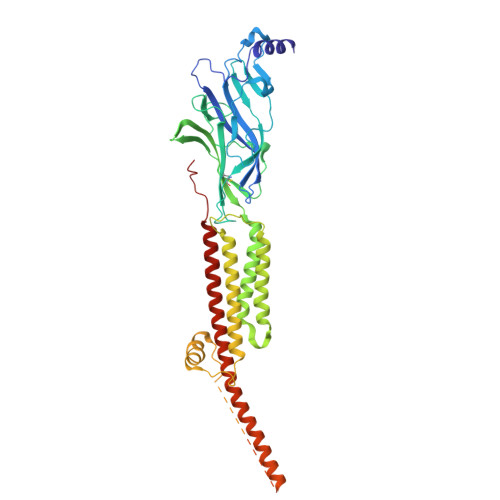 | |
UniProt | |||||
Find proteins for P02712 (Tetronarce californica) Explore P02712 Go to UniProtKB: P02712 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P02712 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Acetylcholine receptor subunit gamma | 489 | Tetronarce californica | Mutation(s): 0 Membrane Entity: Yes | 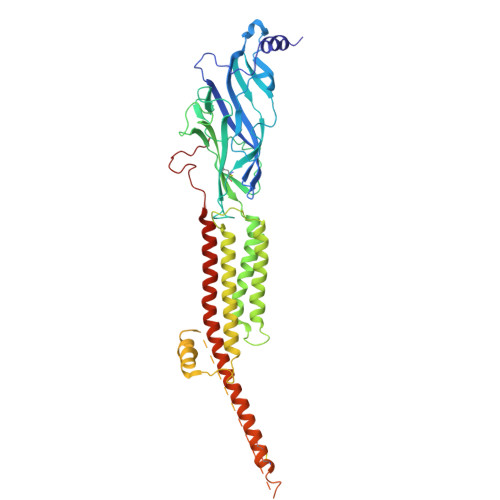 | |
UniProt | |||||
Find proteins for P02714 (Tetronarce californica) Explore P02714 Go to UniProtKB: P02714 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P02714 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Erabutoxin a | 62 | Laticauda semifasciata | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for P60775 (Laticauda semifasciata) Explore P60775 Go to UniProtKB: P60775 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P60775 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | H, I | 9 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G83161QT GlyCosmos: G83161QT GlyGen: G83161QT | |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | J, K | 3 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G15407YE GlyCosmos: G15407YE GlyGen: G15407YE | |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | L | 5 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G08466KH GlyCosmos: G08466KH GlyGen: G08466KH | |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| POV Query on POV | M [auth A], Q [auth C], R [auth C], S [auth D] | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate C42 H82 N O8 P WTJKGGKOPKCXLL-PFDVCBLKSA-N |  | ||
| PLM Query on PLM | U [auth E] | PALMITIC ACID C16 H32 O2 IPCSVZSSVZVIGE-UHFFFAOYSA-N |  | ||
| NAG Query on NAG | N [auth B], O [auth B], T [auth E] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| OCT Query on OCT | P [auth B] | N-OCTANE C8 H18 TVMXDCGIABBOFY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| RECONSTRUCTION | cryoSPARC | 4.4 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Ministry of Education (MoE, Singapore) | Singapore | -- |