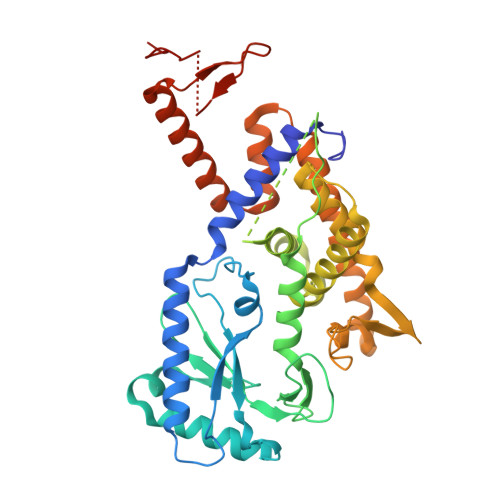
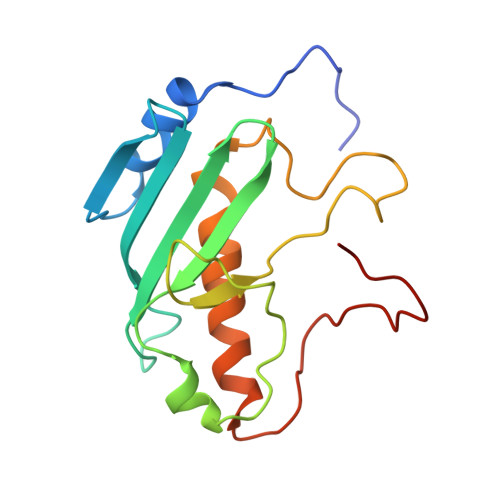
Phage defence system CBASS is regulated by a prokaryotic E2 enzyme that imitates the ubiquitin pathway.
Yan, Y., Xiao, J., Huang, F., Xian, W., Yu, B., Cheng, R., Wu, H., Lu, X., Wang, X., Huang, W., Li, J., Oyejobi, G.K., Robinson, C.V., Wu, H., Wu, D., Liu, X., Wang, L., Zhu, B.(2024) Nat Microbiol 9: 1566-1578
- PubMed: 38649411
- DOI: https://doi.org/10.1038/s41564-024-01684-z
- Primary Citation of Related Structures:
8HSB, 8YJY - PubMed Abstract:
The cyclic-oligonucleotide-based anti-phage signalling system (CBASS) is a type of innate prokaryotic immune system. Composed of a cyclic GMP-AMP synthase (cGAS) and CBASS-associated proteins, CBASS uses cyclic oligonucleotides to activate antiviral immunity. One major class of CBASS contains a homologue of eukaryotic ubiquitin-conjugating enzymes, which is either an E1-E2 fusion or a single E2. However, the functions of single E2s in CBASS remain elusive. Here, using biochemical, genetic, cryo-electron microscopy and mass spectrometry investigations, we discover that the E2 enzyme from Serratia marcescens regulates cGAS by imitating the ubiquitination cascade. This includes the processing of the cGAS C terminus, conjugation of cGAS to a cysteine residue, ligation of cGAS to a lysine residue, cleavage of the isopeptide bond and poly-cGASylation. The poly-cGASylation activates cGAS to produce cGAMP, which acts as an antiviral signal and leads to cell death. Thus, our findings reveal a unique regulatory role of E2 in CBASS.
- Key Laboratory of Molecular Biophysics, the Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan, China.
Organizational Affiliation: