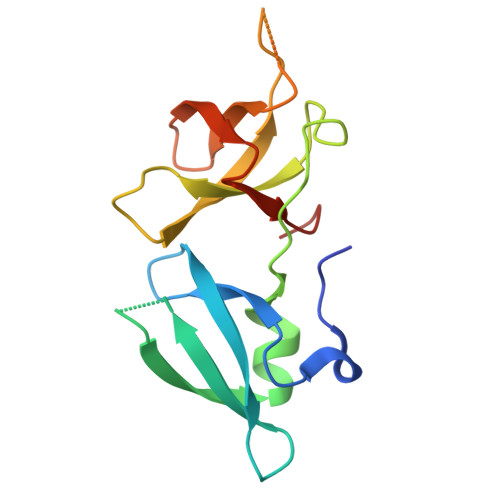
BAHCC1 binds H4K20me1 to facilitate the MCM complex loading and DNA replication.
Li, D., Zhang, Z.M., Mei, L., Yu, Y., Guo, Y., Mackintosh, S.G., Chen, J., Allison, D.F., Kim, A., Storey, A.J., Edmondson, R.D., Byrum, S.D., Tackett, A.J., Cai, L., Cook, J.G., Song, J., Wang, G.G.(2025) Nat Commun 16: 5502-5502
- PubMed: 40592879
- DOI: https://doi.org/10.1038/s41467-025-61284-1
- Primary Citation of Related Structures:
8YE8 - PubMed Abstract:
Mono-methylation of histone H4 lysine 20 (H4K20me1) regulates DNA replication, cell cycle progression and DNA damage repair. How exactly H4K20me1 regulates these biological processes remains unclear. Here, we report that an evolutionarily conserved tandem Tudor domain (TTD) in BAHCC1 (BAHCC1 TTD ) selectively reads H4K20me1 for facilitating replication origin activation and DNA replication. Our biochemical, structural, genomic and cellular analyses demonstrate that BAHCC1 TTD preferentially recognizes H4K20me1 to promote the recruitment of BAHCC1 and its interacting partners, notably Mini-chromosome Maintenance (MCM) complex, to replication origin sites. Combined actions of the H4K20me1-reading BAHCC1 and the H4K20me2-reading Origin Recognition Complex (ORC) ensure genomic loading of MCM for replication. Depletion of BAHCC1, or disruption of the BAHCC1 TTD :H4K20me1 interaction, reduces H4K20me1 levels and MCM loading, leading to defects in replication origin activation and cell cycle progression. In summary, this study identifies BAHCC1 TTD as an effector transducing H4K20me1 signals into MCM recruitment to promote DNA replication.
- Department of Biochemistry and Biophysics, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27599, USA.
Organizational Affiliation: