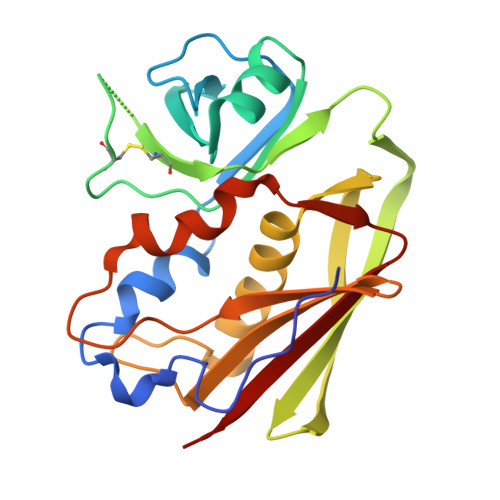
Structural insights into the binding of nanobodies to the Staphylococcal enterotoxin B.
Zong, X., Liu, P., Wang, Z., Zhu, H., Zhong, C., Zhong, P., Jiang, H., Liu, J., Ma, Z., Liu, X., Liu, R., Ding, Y.(2024) Int J Biol Macromol 276: 133957-133957
- PubMed: 39029852
- DOI: https://doi.org/10.1016/j.ijbiomac.2024.133957
- Primary Citation of Related Structures:
8YBL, 8YBM, 8YBN, 8YBO, 8YBP - PubMed Abstract:
Staphylococcal Enterotoxin Type B (SEB), produced by Staphylococcus aureus bacteria, is notorious for inducing severe food poisoning and toxic shock syndrome. While nanobody-based treatments hold promises for combating SEB-induced diseases, the lack of structural information between SEB and nanobodies has hindered the development of nanobody-based therapeutics. Here, we present crystal structures of SEB-Nb3, SEB-Nb6, SEB-Nb8, SEB-Nb11, and SEB-Nb20 at resolutions ranging from 1.59 Å to 2.33 Å. Crystallographic analysis revealed that Nb3, Nb8, Nb11, and Nb20 bind to SEB at the T-cell receptor (TCR) interface, while Nb6 binds at the major histocompatibility complex (MHC) interface, suggesting their potential to inhibit SEB function by disrupting interactions with TCR or MHC molecules. Molecular biological analyses confirmed the thermodynamic and kinetic parameters of Nb3, Nb5, Nb6, Nb8, Nb11, Nb15, Nb18, and Nb20 to SEB. The competitive inhibition was further confirmed by cell-based experiments demonstrating nanobody neutralization. These findings elucidate the structural basis for developing specific nanobodies to neutralize SEB threats, providing crucial insights into the underlying mechanisms and offering significant assistance for further optimization towards future therapeutic strategies.
- State Key Laboratory of Genetic Engineering, School of Life Sciences, Fudan University, Shanghai 200433, China.
Organizational Affiliation: