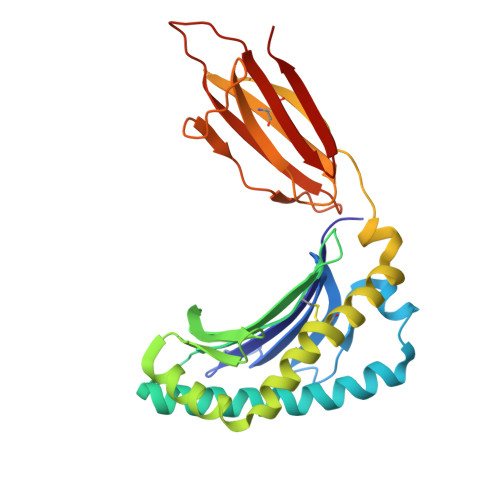
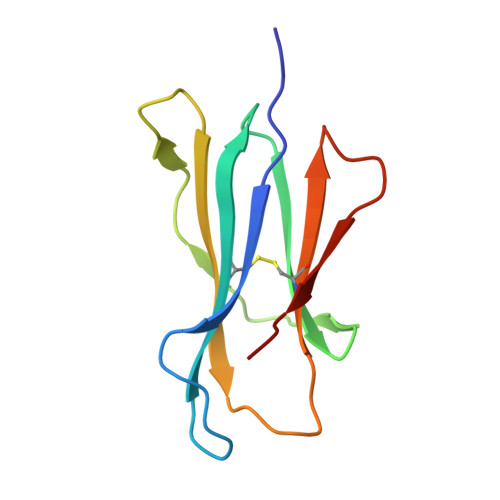
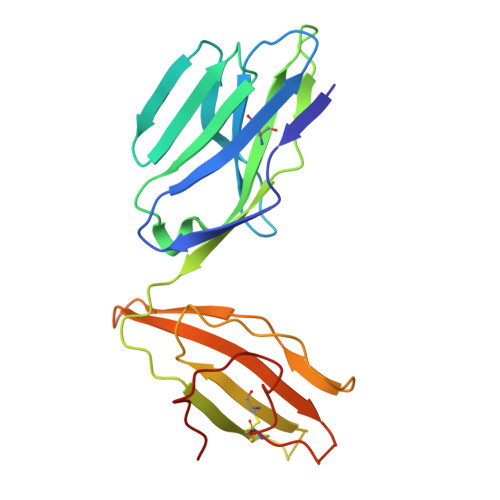
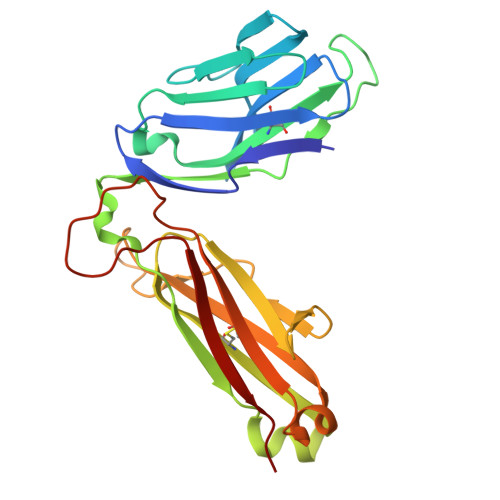
Development of Ribityllumazine Analogue as Mucosal-Associated Invariant T Cell Ligands.
Takasaki, R., Ito, E., Nagae, M., Takahashi, Y., Matsuoka, T., Yasue, W., Arichi, N., Ohno, H., Yamasaki, S., Inuki, S.(2024) J Am Chem Soc 146: 29964-29976
- PubMed: 39432319
- DOI: https://doi.org/10.1021/jacs.4c12997
- Primary Citation of Related Structures:
8Y6X - PubMed Abstract:
Mucosal-associated invariant T (MAIT) cells are a subset of innate-like T cells abundant in human tissues that play a significant role in defense against bacterial and viral infections and in tissue repair. MAIT cells are activated by recognizing microbial-derived small-molecule ligands presented by the MHC class I related-1 protein. Although several MAIT cell modulators have been identified in the past decade, potent and chemically stable ligands remain limited. Herein, we carried out a structure-activity relationship study of ribityllumazine derivatives and found a chemically stable MAIT cell ligand with a pteridine core and a 2-oxopropyl group as the Lys-reactive group. The ligand showed high potency in a cocultivation assay using model cell lines of antigen-presenting cells and MAIT cells. The X-ray crystallographic analysis revealed the binding mode of the ligand to MR1 and the T cell receptor, indicating that it forms a covalent bond with MR1 via Schiff base formation. Furthermore, we found that the ligand stimulated proliferation of human MAIT cells in human peripheral blood mononuclear cells and showed an adjuvant effect in mice. Our developed ligand is one of the most potent among chemically stable MAIT cell ligands, contributing to accelerating therapeutic applications of MAIT cells.
- Graduate School of Pharmaceutical Sciences, Kyoto University, Kyoto, Kyoto 606-8501, Japan.
Organizational Affiliation: