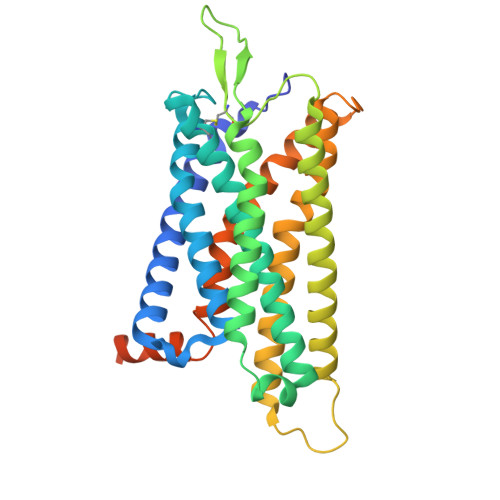
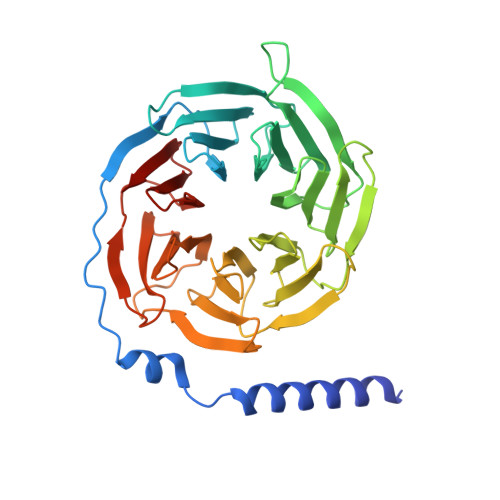
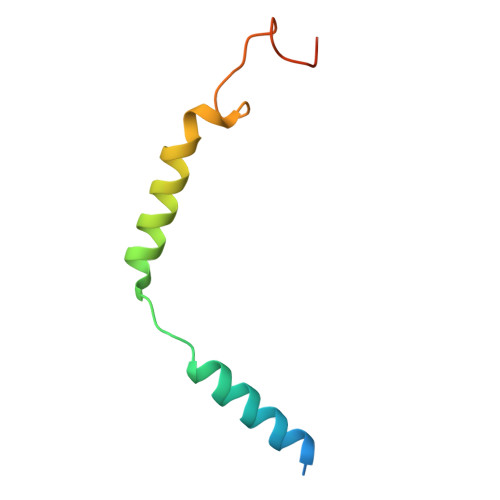
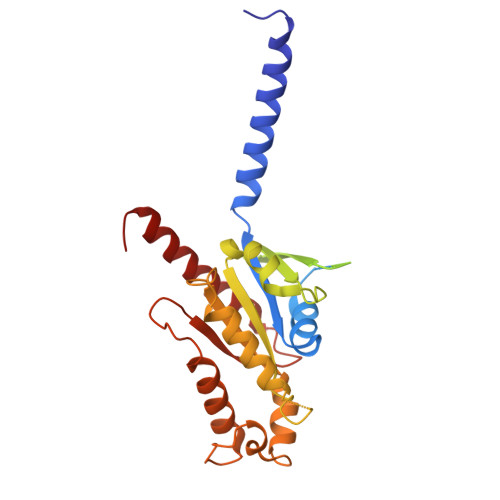
Molecular basis for the activation of PAF receptor by PAF.
Fan, W., Xu, Y., He, X., Luo, P., Zhu, J., Li, J., Wang, R., Yuan, Q., Wu, K., Hu, W., Zhao, Y., Xu, S., Cheng, X., Wang, Y., Xu, H.E., Zhuang, Y.(2024) Cell Rep 43: 114422-114422
- PubMed: 38943642
- DOI: https://doi.org/10.1016/j.celrep.2024.114422
- Primary Citation of Related Structures:
8XYD - PubMed Abstract:
Platelet-activating factor (PAF) is a potent phospholipid mediator crucial in multiple inflammatory and immune responses through binding and activating the PAF receptor (PAFR). However, drug development targeting the PAFR has been limited, partly due to an incomplete understanding of its activation mechanism. Here, we present a 2.9-Å structure of the PAF-bound PAFR-G i complex. Structural and mutagenesis analyses unveil a specific binding mode of PAF, with the choline head forming cation-π interactions within PAFR hydrophobic pocket, while the alkyl tail penetrates deeply into an aromatic cleft between TM4 and TM5. Binding of PAF modulates conformational changes in key motifs of PAFR, triggering the outward movement of TM6, TM7, and helix 8 for G protein coupling. Molecular dynamics simulation suggests a membrane-side pathway for PAF entry into PAFR via the TM4-TM5 cavity. By providing molecular insights into PAFR signaling, this work contributes a foundation for developing therapeutic interventions targeting PAF signal axis.
- School of Chinese Materia Medica, Nanjing University of Chinese Medicine, Nanjing 210046, China; The State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China.
Organizational Affiliation: