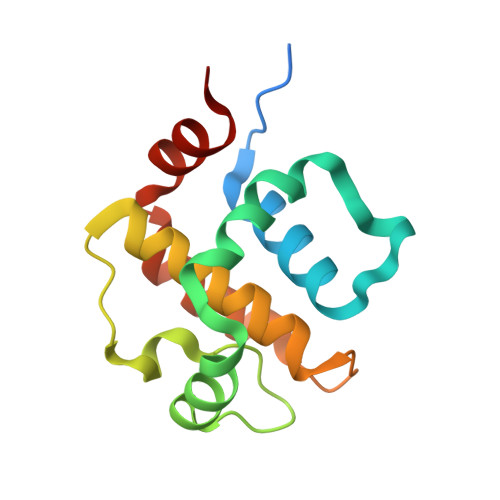
Structural insights into MPXV P1 protein and its orthologs reveal conformational dynamics and a conserved antiviral pocket.
Ni, X., Wang, Y., Fan, X., Yu, J., Lei, J.(2025) Emerg Microbes Infect 14: 2515408-2515408
- PubMed: 40476512
- DOI: https://doi.org/10.1080/22221751.2025.2515408
- Primary Citation of Related Structures:
8XY1, 8XY2, 8XY3, 8XY4 - National Clinical Research Center for Geriatrics, and State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, Chengdu, Sichuan, China.
Organizational Affiliation: