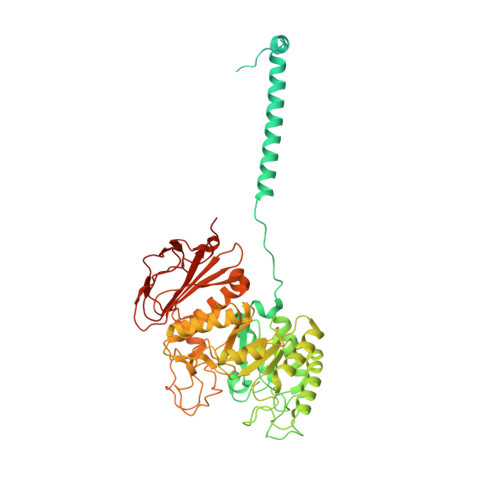
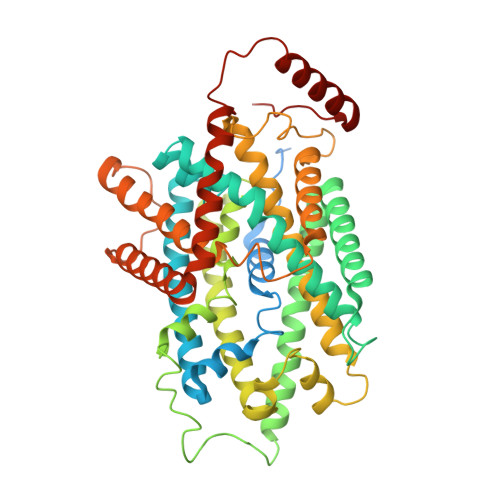
Structural basis for the inhibition mechanism of LAT1-4F2hc complex by JPH203.
Hu, Z., Yan, R.(2024) Cell Discov 10: 73-73
- PubMed: 38956038
- DOI: https://doi.org/10.1038/s41421-024-00697-6
- Primary Citation of Related Structures:
8XPU - Department of Biochemistry, School of Medicine, Key University Laboratory of Metabolism and Health of Guangdong, Institute for Biological Electron Microscopy, Southern University of Science and Technology, Shenzhen, Guangdong, China.
Organizational Affiliation: