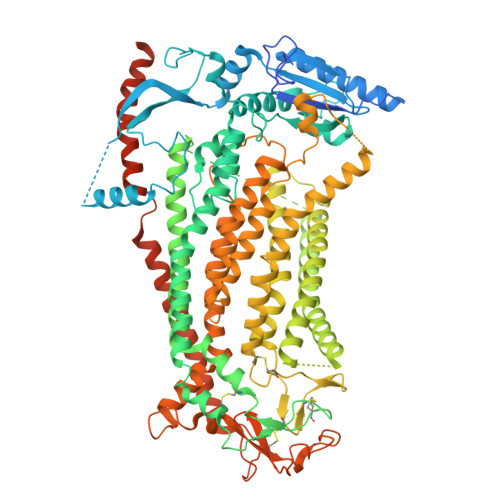
Tamsulosin ameliorates bone loss by inhibiting the release of Cl - through wedging into an allosteric site of TMEM16A.
Li, S., Sun, W., Li, S., Zhu, L., Guo, S., He, J., Li, Y., Tian, C., Zhao, Z., Yu, T., Li, J., Zhang, Y., Hai, Y., Wang, J., Zheng, Y., Wang, R., Hu, X., Ling, S., Li, H., Li, Y.(2025) Proc Natl Acad Sci U S A 122: e2407493121-e2407493121
- PubMed: 39739807
- DOI: https://doi.org/10.1073/pnas.2407493121
- Primary Citation of Related Structures:
8XLR - PubMed Abstract:
TMEM16A, a key calcium-activated chloride channel, is crucial for many physiological and pathological processes such as cancer, hypertension, and osteoporosis, etc. However, the regulatory mechanism of TMEM16A is poorly understood, limiting the discovery of effective modulators. Here, we unveil an allosteric gating mechanism by presenting a high-resolution cryo-EM structure of TMEM16A in complex with a channel inhibitor that we identified, Tamsulosin, which is resolved at 2.93 Å. Tamsulosin wedges itself into a pocket within the extracellular domain of TMEM16A, surrounded by α1-α2, α5-α6, and α9-α10 loops. This binding stabilizes a transient preopen conformation of TMEM16A, which is activated by Ca 2+ ions while still preserving a closed pore to prevent Cl - permeation. Validation of this binding site through computational, electrophysiological, and functional experiments, along with site-directed mutagenesis, confirmed the pivotal roles of the pocket-lining residues R605 and E624 on α5-α6 loop in modulating Tamsulosin binding and pore activity. Tamsulosin induces significant positional shifts in extracellular loops, particularly the α5-α6 loop, which moves toward the extracellular exit of the pore, leading to noticeable structural rearrangements in pore-lining helices. The hinges induced by P595 in α5 and G711 in α7 introduce flexibility to the transmembrane helices, orienting Y593 to collaborate with I641 in effectively gating the preopening pore. Notably, Tamsulosin demonstrates significant antiosteoporotic effects by inhibiting TMEM16A, suggesting potential for its repurposing in new therapeutic indications. Our study not only enhances our understanding of the gating mechanism of TMEM16A inhibition but also facilitates structure-based drug design targeting TMEM16A.
- Innovation Center for AI and Drug Discovery, School of Pharmacy, East China Normal University, Shanghai 200062, China.
Organizational Affiliation: