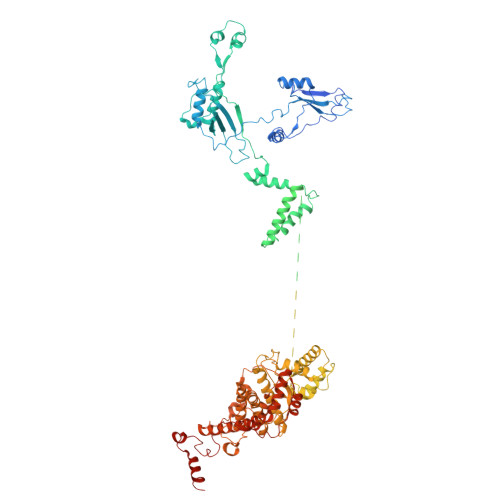
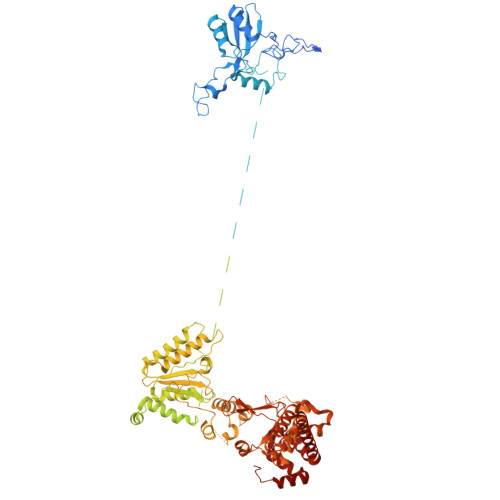
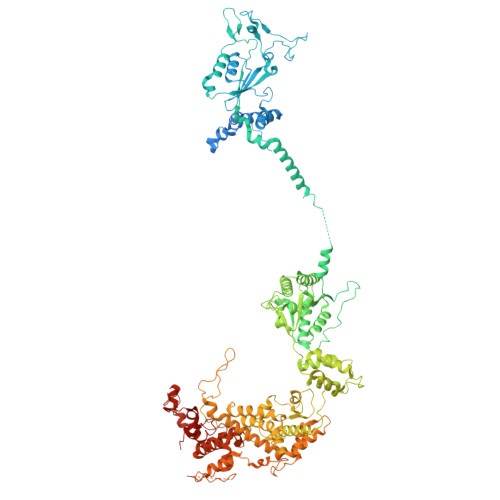
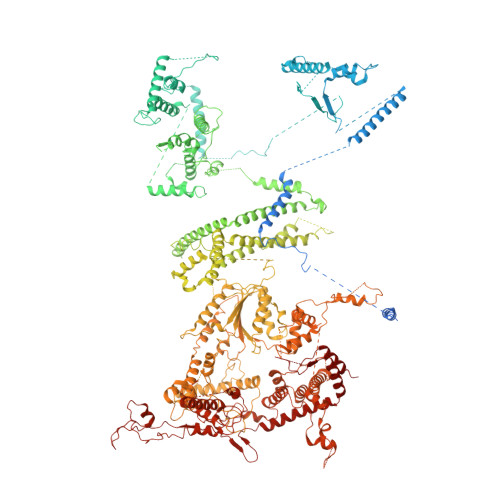
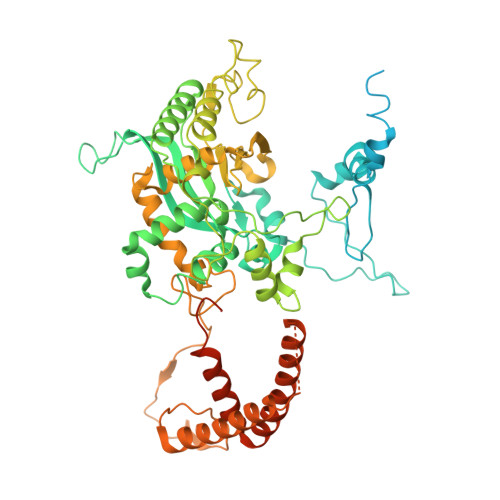
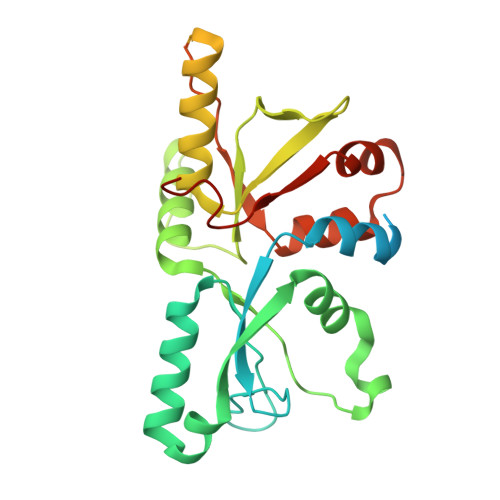
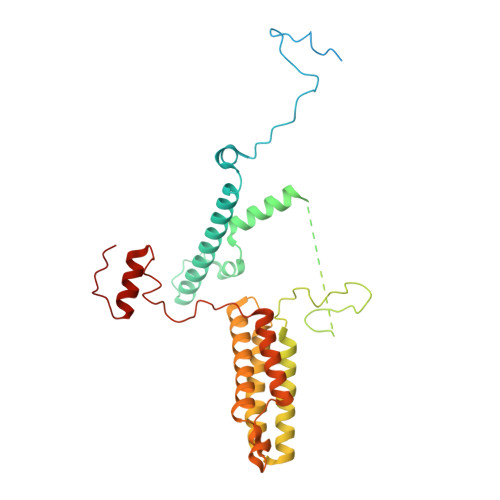
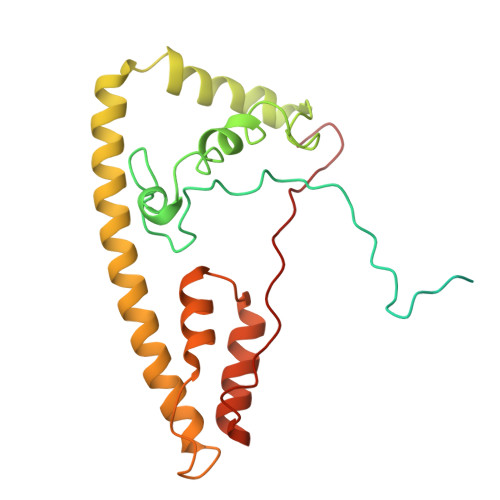
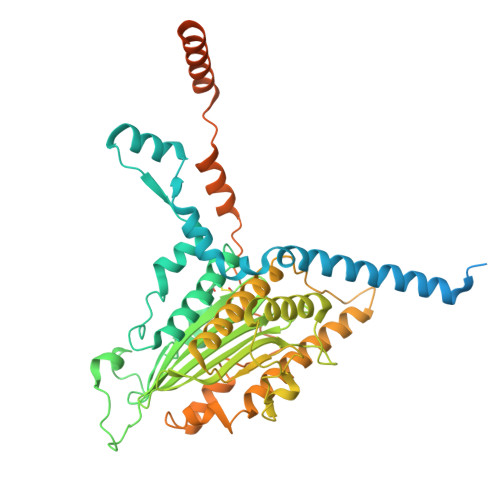
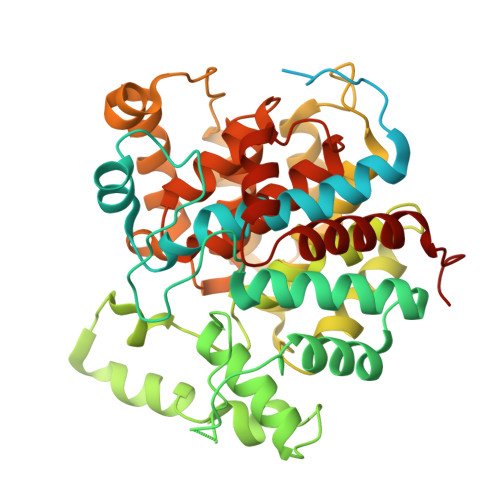
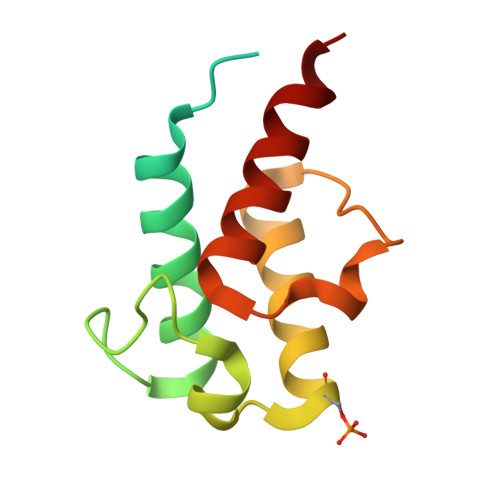
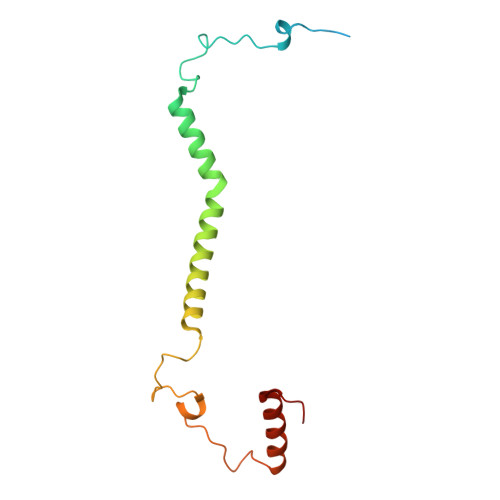
Architecture of the ATP-driven motor for protein import into chloroplasts.
Wang, N., Xing, J., Su, X., Pan, J., Chen, H., Shi, L., Si, L., Yang, W., Li, M.(2024) Mol Plant 17: 1702-1718
- PubMed: 39327731
- DOI: https://doi.org/10.1016/j.molp.2024.09.010
- Primary Citation of Related Structures:
8XKS - PubMed Abstract:
Thousands of nuclear-encoded proteins are transported into chloroplasts through the TOC-TIC translocon that spans the chloroplast envelope membranes. A motor complex pulls the translocated proteins out of the TOC-TIC complex into the chloroplast stroma by hydrolyzing ATP. The Orf2971-FtsHi complex has been suggested to serve as the ATP-hydrolyzing motor in Chlamydomonas reinhardtii, but little is known about its architecture and assembly. Here, we report the 3.2-Å resolution structure of the Chlamydomonas Orf2971-FtsHi complex. The 20-subunit complex spans the chloroplast inner envelope, with two bulky modules protruding into the intermembrane space and stromal matrix. Six subunits form a hetero-hexamer that potentially provides the pulling force through ATP hydrolysis. The remaining subunits, including potential enzymes/chaperones, likely facilitate the complex assembly and regulate its proper function. Taken together, our results provide the structural foundation for a mechanistic understanding of chloroplast protein translocation.
- Key Laboratory of Biomacromolecules (CAS), National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: