Structure of G protein-coupled receptor GPR1 bound to full-length chemerin adipokine reveals a chemokine-like reverse binding mode.
Liu, A., Liu, Y., Chen, G., Lyu, W., Ye, F., Wang, J., Liao, Q., Zhu, L., Du, Y., Ye, R.D.(2024) PLoS Biol 22: e3002838-e3002838
- PubMed: 39466725
- DOI: https://doi.org/10.1371/journal.pbio.3002838
- Primary Citation of Related Structures:
8JJP, 8XGM - PubMed Abstract:
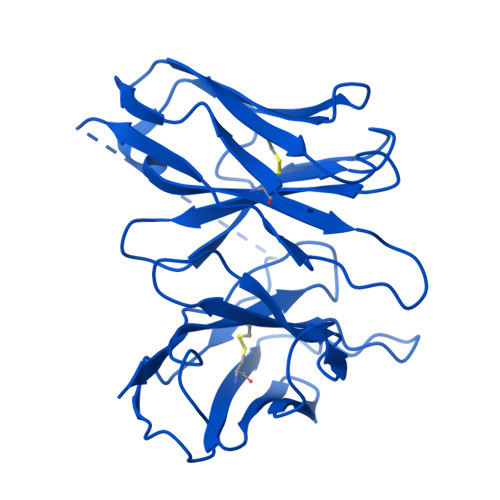
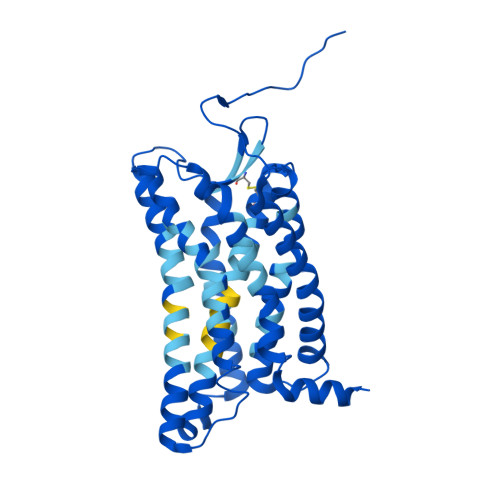
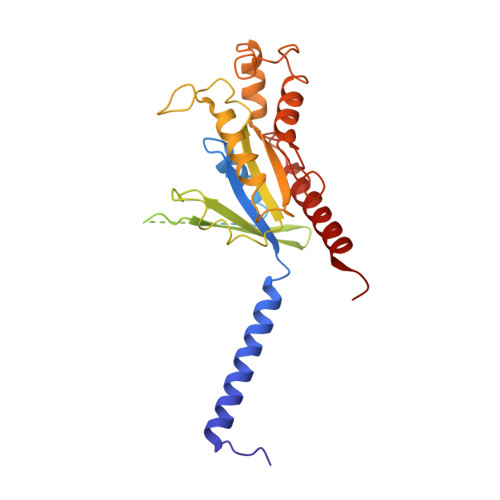
Chemerin is an adipokine with chemotactic activity to a subset of leukocytes. Chemerin binds to 3 G protein-coupled receptors, including chemokine-like receptor 1 (CMKLR1), G protein-coupled receptor 1 (GPR1), and C-C chemokine receptor-like 2 (CCRL2). Here, we report that GPR1 is capable of Gi signaling when stimulated with full-length chemerin or its C-terminal nonapeptide (C9, YFPGQFAFS). We present high-resolution cryo-EM structures of Gi-coupled GPR1 bound to full-length chemerin and to the C9 peptide, respectively. C9 insertion into the transmembrane (TM) binding pocket is both necessary and sufficient for GPR1 signaling, whereas the full-length chemerin uses its bulky N-terminal core for interaction with a β-strand located at the N-terminus of GPR1. This interaction involves multiple β-strands of full-length chemerin, forming a β-sheet that serves as a "lid" for the TM binding pocket and is energetically expensive to remove as indicated by molecular dynamics simulations with free energy landscape analysis. Combining results from functional assays, our structural model explains why C9 is an activating peptide at GPR1 and how the full-length chemerin uses a "two-site" model for enhanced interaction with GPR1.
- Kobilka Institute of Innovative Drug Discovery, School of Medicine, The Chinese University of Hong Kong, Shenzhen, China.
Organizational Affiliation: