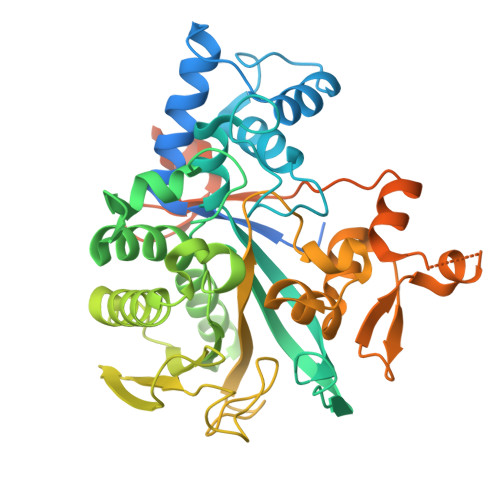
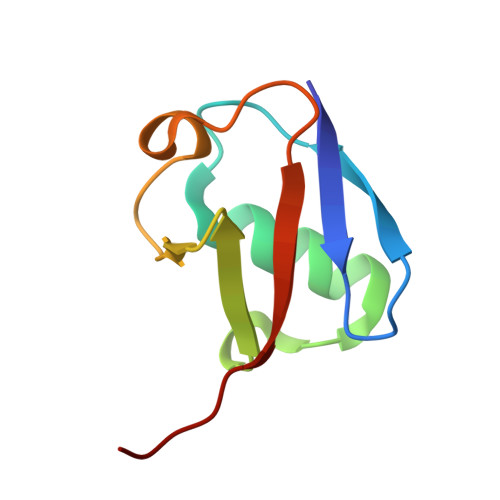
Legionella effector LnaB is a phosphoryl AMPylase that impairs phosphosignalling.
Wang, T., Song, X., Tan, J., Xian, W., Zhou, X., Yu, M., Wang, X., Xu, Y., Wu, T., Yuan, K., Ran, Y., Yang, B., Fan, G., Liu, X., Zhou, Y., Zhu, Y.(2024) Nature 631: 393-401
- PubMed: 38776962
- DOI: https://doi.org/10.1038/s41586-024-07573-z
- Primary Citation of Related Structures:
8JO3, 8JO4, 8XEP - PubMed Abstract:
AMPylation is a post-translational modification in which AMP is added to the amino acid side chains of proteins 1,2 . Here we show that, with ATP as the ligand and actin as the host activator, the effector protein LnaB of Legionella pneumophila exhibits AMPylase activity towards the phosphoryl group of phosphoribose on PR R42 -Ub that is generated by the SidE family of effectors, and deubiquitinases DupA and DupB in an E1- and E2-independent ubiquitination process 3-7 . The product of LnaB is further hydrolysed by an ADP-ribosylhydrolase, MavL, to Ub, thereby preventing the accumulation of PR R42 -Ub and ADPR R42 -Ub and protecting canonical ubiquitination in host cells. LnaB represents a large family of AMPylases that adopt a common structural fold, distinct from those of the previously known AMPylases, and LnaB homologues are found in more than 20 species of bacterial pathogens. Moreover, LnaB also exhibits robust phosphoryl AMPylase activity towards phosphorylated residues and produces unique ADPylation modifications in proteins. During infection, LnaB AMPylates the conserved phosphorylated tyrosine residues in the activation loop of the Src family of kinases 8,9 , which dampens downstream phosphorylation signalling in the host. Structural studies reveal the actin-dependent activation and catalytic mechanisms of the LnaB family of AMPylases. This study identifies, to our knowledge, an unprecedented molecular regulation mechanism in bacterial pathogenesis and protein phosphorylation.
- Department of Gastroenterology of the Second Affiliated Hospital, School of Medicine and College of Animal Sciences, Life Sciences Institute, Zhejiang University, Hangzhou, China.
Organizational Affiliation: