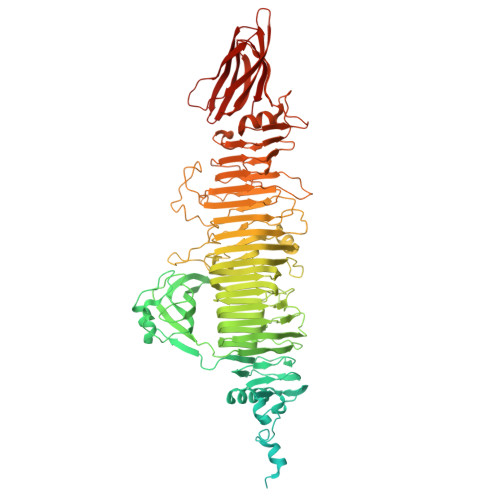
Structural and functional basis of bacteriophage K64-ORF41 depolymerase for capsular polysaccharide degradation of Klebsiella pneumoniae K64.
Huang, T., Zhang, Z., Tao, X., Shi, X., Lin, P., Liao, D., Ma, C., Cai, X., Lin, W., Jiang, X., Luo, P., Wu, S., Xie, Y.(2024) Int J Biol Macromol 265: 130917-130917
- PubMed: 38513899
- DOI: https://doi.org/10.1016/j.ijbiomac.2024.130917
- Primary Citation of Related Structures:
8X8M, 8X8O - PubMed Abstract:
Capsule polysaccharide is an important virulence factor of Klebsiella pneumoniae (K. pneumoniae), which protects bacteria against the host immune response. A promising therapeutic approach is using phage-derived depolymerases to degrade the capsular polysaccharide and expose and sensitize the bacteria to the host immune system. Here we determined the cryo-electron microscopy (cryo-EM) structures of a bacteriophage tail-spike protein against K. pneumoniae K64, ORF41 (K64-ORF41) and ORF41 in EDTA condition (K64-ORF41 EDTA ), at 2.37 Å and 2.50 Å resolution, respectively, for the first time. K64-ORF41 exists as a trimer and each protomer contains a β-helix domain including a right-handed parallel β-sheet helix fold capped at both ends, an insertion domain, and one β-sheet jellyroll domain. Moreover, our structural comparison with other depolymerases of K. pneumoniae suggests that the catalytic residues (Tyr528, His574 and Arg628) are highly conserved although the substrate of capsule polysaccharide is variable. Besides that, we figured out the important residues involved in the substrate binding pocket including Arg405, Tyr526, Trp550 and Phe669. This study establishes the structural and functional basis for the promising phage-derived broad-spectrum activity depolymerase therapeutics and effective CPS-degrading agents for the treatment of carbapenem-resistant K. pneumoniae K64 infections.
- Department of Neurosurgery, Xijing Hospital, Fourth Military Medical University, Xi'an, Shaanxi 710032, China; Department of Pharmacy, Xi'an No.3 Hospital, the Affiliated Hospital of Northwest University, Xi'an, Shaanxi 710021, China; School of Pharmacy, Health Science Center, Xi'an Jiaotong University, Xi'an, Shaanxi 710061, China.
Organizational Affiliation: