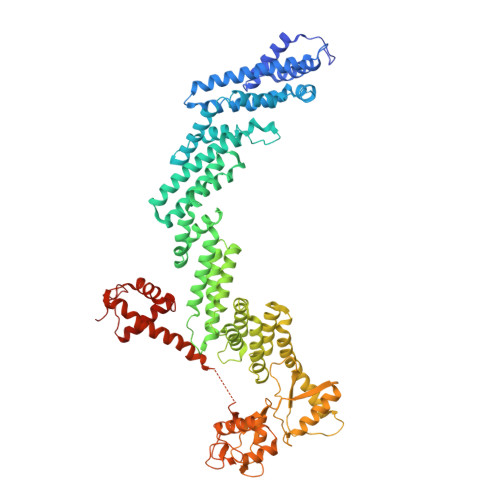
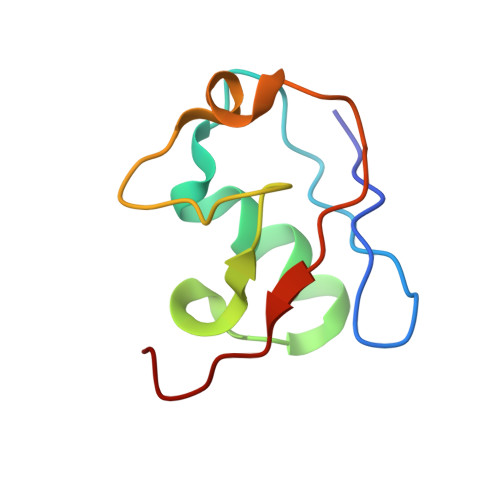
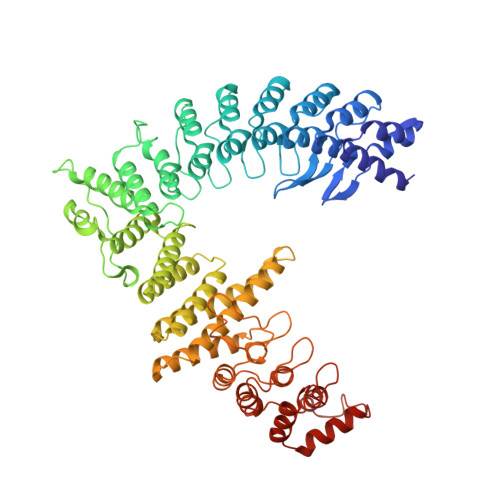
Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Chen, X., Raiff, A., Li, S., Guo, Q., Zhang, J., Zhou, H., Timms, R.T., Yao, X., Elledge, S.J., Koren, I., Zhang, K., Xu, C.(2024) Nat Commun 15: 3558-3558
- PubMed: 38670995
- DOI: https://doi.org/10.1038/s41467-024-47890-5
- Primary Citation of Related Structures:
8WQA, 8WQB, 8WQC, 8WQD, 8WQE, 8WQF, 8WQG, 8WQH, 8WQI - PubMed Abstract:
The E3 ligase-degron interaction determines the specificity of the ubiquitin‒proteasome system. We recently discovered that FEM1B, a substrate receptor of Cullin 2-RING ligase (CRL2), recognizes C-degrons containing a C-terminal proline. By solving several cryo-EM structures of CRL2 FEM1B bound to different C-degrons, we elucidate the dimeric assembly of the complex. Furthermore, we reveal distinct dimerization states of unmodified and neddylated CRL2 FEM1B to uncover the NEDD8-mediated activation mechanism of CRL2 FEM1B . Our research also indicates that, FEM1B utilizes a bipartite mechanism to recognize both the C-terminal proline and an upstream aromatic residue within the substrate. These structural findings, complemented by in vitro ubiquitination and in vivo cell-based assays, demonstrate that CRL2 FEM1B -mediated polyubiquitination and subsequent protein turnover depend on both FEM1B-degron interactions and the dimerization state of the E3 ligase complex. Overall, this study deepens our molecular understanding of how Cullin-RING E3 ligase substrate selection mediates protein turnover.
- MOE Key Laboratory for Cellular Dynamics, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, 230027, PR China.
Organizational Affiliation: