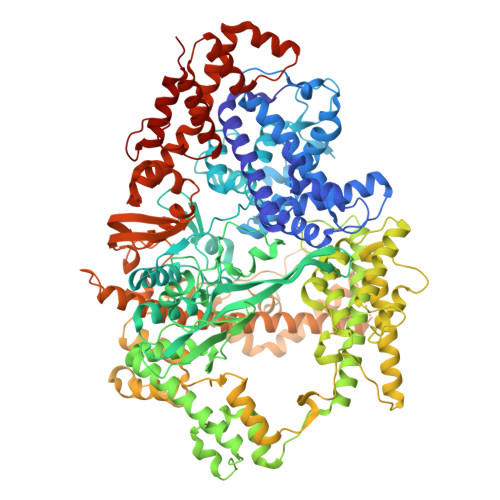
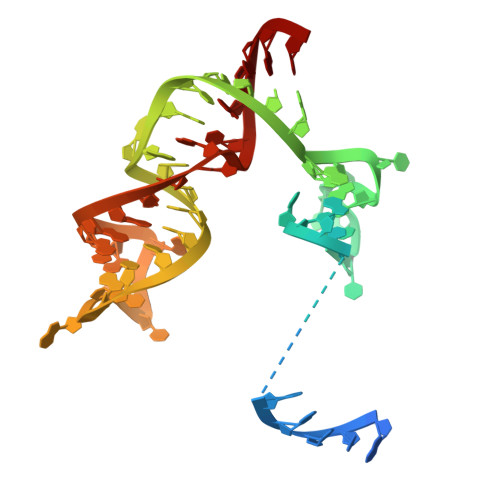
Structural insights into RNA-guided RNA editing by the Cas13b-ADAR2 complex.
Ishikawa, J., Kato, K., Kannan, S., Okazaki, S., Ishiguro, S., Yamashita, K., Yachie, N., Nishizawa, T., Zhang, F., Nishimasu, H.(2025) Nat Struct Mol Biol 32: 1567-1578
- PubMed: 40217120
- DOI: https://doi.org/10.1038/s41594-025-01529-1
- Primary Citation of Related Structures:
8WF8, 8WF9, 8WFA, 8WFB - PubMed Abstract:
Cas13 is an RNA-guided RNA endonuclease derived from the type VI CRISPR-Cas system, which has been used in numerous RNA-targeting technologies, such as RNA knockdown, detection and editing. The catalytically inactive Prevotella sp. Cas13b (dPspCas13b) fused to the human adenosine deaminase acting on RNA 2 (ADAR2) deaminase domain can edit adenosine in target transcripts to inosine, in an RNA-editing technology called REPAIR (RNA editing for programmable A-to-I replacement), which has potential for gene therapy. Here we report the cryo-electron microscopy structures of the PspCas13b-guide RNA binary complex, the PspCas13b-guide RNA-target RNA ternary complex and the dPspCas13b-ADAR2-guide RNA-target RNA complex. These structures provide mechanistic insights into RNA cleavage and editing. We applied our structural insights to engineer a compact and efficient dPspCas13b-ADAR2 complex (REPAIR-mini). Overall, our findings advance the understanding of CRISPR-Cas13 effector nucleases and could enable the development of improved RNA-targeting technologies.
- Department of Chemistry and Biotechnology, Graduate School of Engineering, The University of Tokyo, Tokyo, Japan.
Organizational Affiliation: