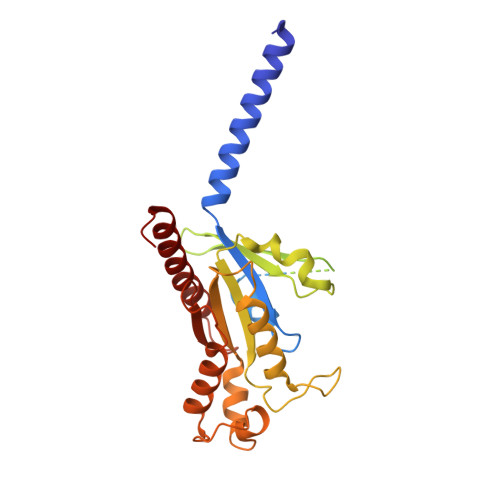
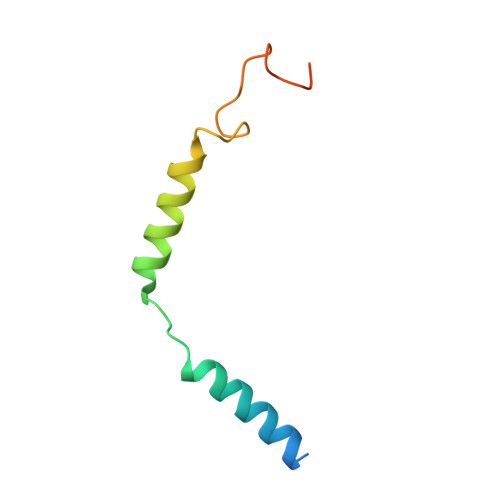
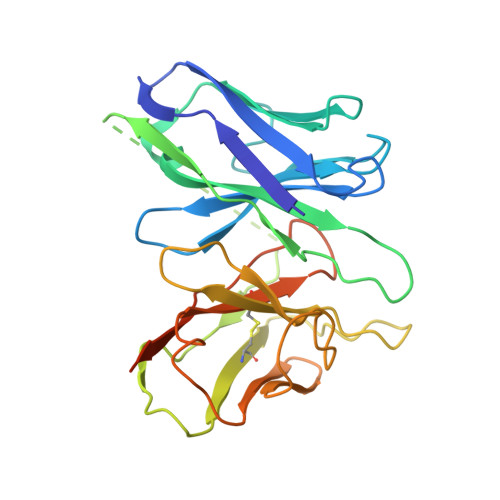
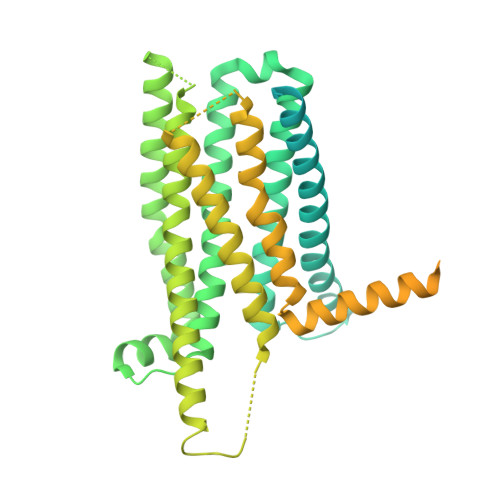
Bitter taste receptor activation by cholesterol and an intracellular tastant.
Kim, Y., Gumpper, R.H., Liu, Y., Kocak, D.D., Xiong, Y., Cao, C., Deng, Z., Krumm, B.E., Jain, M.K., Zhang, S., Jin, J., Roth, B.L.(2024) Nature 628: 664-671
- PubMed: 38600377
- DOI: https://doi.org/10.1038/s41586-024-07253-y
- Primary Citation of Related Structures:
8VY7, 8VY9 - PubMed Abstract:
Bitter taste sensing is mediated by type 2 taste receptors (TAS2Rs (also known as T2Rs)), which represent a distinct class of G-protein-coupled receptors 1 . Among the 26 members of the TAS2Rs, TAS2R14 is highly expressed in extraoral tissues and mediates the responses to more than 100 structurally diverse tastants 2-6 , although the molecular mechanisms for recognizing diverse chemicals and initiating cellular signalling are still poorly understood. Here we report two cryo-electron microscopy structures for TAS2R14 complexed with G gust (also known as gustducin) and G i1 . Both structures have an orthosteric binding pocket occupied by endogenous cholesterol as well as an intracellular allosteric site bound by the bitter tastant cmpd28.1, including a direct interaction with the α5 helix of G gust and G i1 . Computational and biochemical studies validate both ligand interactions. Our functional analysis identified cholesterol as an orthosteric agonist and the bitter tastant cmpd28.1 as a positive allosteric modulator with direct agonist activity at TAS2R14. Moreover, the orthosteric pocket is connected to the allosteric site via an elongated cavity, which has a hydrophobic core rich in aromatic residues. Our findings provide insights into the ligand recognition of bitter taste receptors and suggest activities of TAS2R14 beyond bitter taste perception via intracellular allosteric tastants.
- Department of Pharmacology, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Organizational Affiliation: