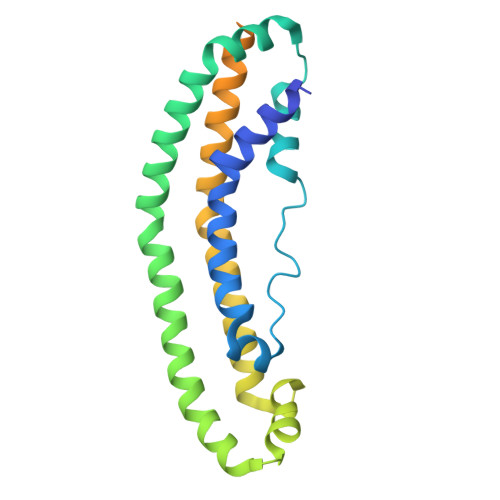
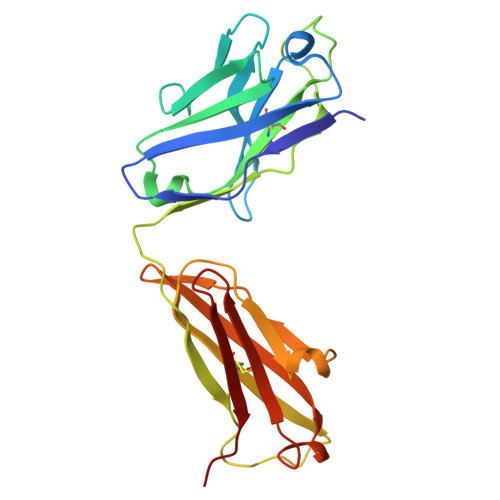
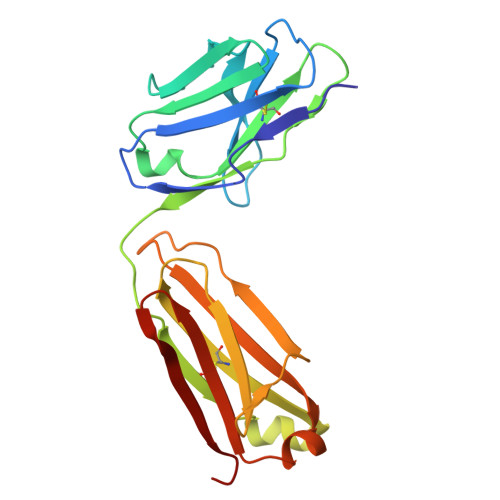
The Structure of the Apolipoprotein A-I Monomer Provides Insights Into Its Oligomerisation and Lipid-binding Mechanisms.
Tou, H.I., Rosenes, Z., Khandokar, Y., Zlatic, C.O., Metcalfe, R.D., Mok, Y.F., Morton, C.J., Gooley, P.R., Griffin, M.D.W.(2025) J Mol Biology 437: 169394-169394
- PubMed: 40816717
- DOI: https://doi.org/10.1016/j.jmb.2025.169394
- Primary Citation of Related Structures:
8VXJ - PubMed Abstract:
Apolipoprotein A-I (apoA-I) plays important roles in clearing cholesterol and phospholipids from peripheral tissues, forming high-density lipoprotein (HDL). However, despite this important function, apoA-I has a propensity to form amyloid fibrils implicated in atherosclerosis and hereditary amyloidosis. Historically, structural determination of lipid-free or lipid-poor apoA-I has been difficult. Here, we obtained the crystal structure of the apoA-I monomer in complex with the antigen-binding fragment (Fab) of a monoclonal antibody. The structure reveals that the N-terminal domain (NTD, residues 1-184) of apoA-I is a compact four-helical bundle, whereas the C-terminal domain (CTD, residues 185-243) is unresolved in the structure. Molecular Dynamics (MD) simulations and small-angle X-ray scattering (SAXS) analysis revealed that the apoA-I NTD dimerises by domain-swapping and the dimer is elongated. Methionine (Met) oxidation in apoA-I destabilises both full-length apoA-I (apoA-I FL ) and C-terminally truncated apoA-I (apoA-I Δ185-243 ), causing dissociation of the domain-swapped dimer and fibril formation. Met oxidation also increased the lipid-binding ability of apoA-I Δ185-243 , while the amyloidogenic mutation, G26R, did not. Hydrogen-deuterium exchange coupled with nuclear magnetic resonance (HDX-NMR), SAXS, and MD analyses showed that triply Met-oxidised (3MetO) and G26R apoA-I Δ185-243 are both highly dynamic but remain partially folded. Based on these results, we propose that domain-swapping dimerisation also exists in apoA-I FL , with the CTD mediating further oligomerisation. We also propose that lipid-binding is promoted by increased global destabilisation in the protein structure, and/or driven by a specific local conformation that is induced by Met-oxidation but not the G26R mutation.
- Department of Biochemistry and Pharmacology, Bio21 Molecular Science and Biotechnology Institute, University of Melbourne, Parkville, Victoria 3010, Australia.
Organizational Affiliation: