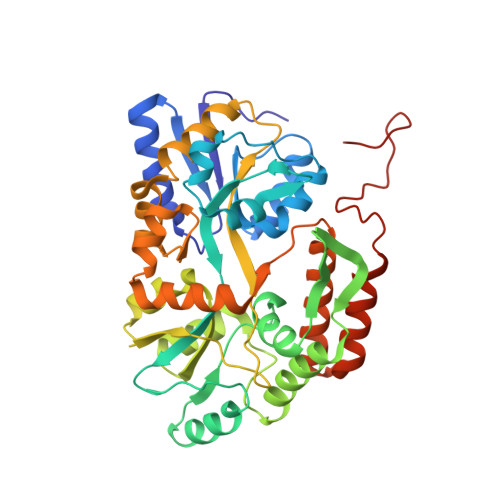
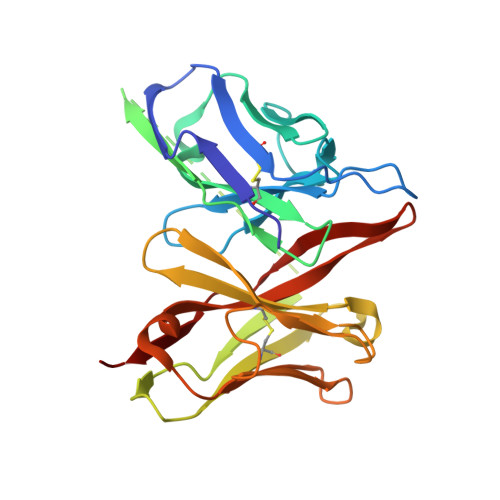
Structural basis for antibody recognition of the proximal MUC16 ectodomain.
Lee, K., Perry, K., Xu, M., Veillard, I., Kumar, R., Rao, T.D., Rueda, B.R., Spriggs, D.R., Yeku, O.O.(2024) J Ovarian Res 17: 41-41
- PubMed: 38374055
- DOI: https://doi.org/10.1186/s13048-024-01373-9
- Primary Citation of Related Structures:
8VRR, 8VRS - PubMed Abstract:
Mucin 16 (MUC16) overexpression is linked with cancer progression, metastasis, and therapy resistance in high grade serous ovarian cancer and other malignancies. The cleavage of MUC16 forms independent bimodular fragments, the shed tandem repeat sequence which circulates as a protein bearing the ovarian cancer biomarker (CA125) and a proximal membrane-bound component which is critical in MUC16 oncogenic behavior. A humanized, high affinity antibody targeting the proximal ectodomain represents a potential therapeutic agent against MUC16 with lower antigenic potential and restricted human tissue expression. Here, we demonstrate the potential therapeutic versatility of the humanized antibody as a monoclonal antibody, antibody drug conjugate, and chimeric antigen receptor. We report the crystal structures of 4H11-scFv, derived from an antibody specifically targeting the MUC16 C-terminal region, alone and in complex with a 26-amino acid MUC16 segment resolved at 2.36 Å and 2.47 Å resolution, respectively. The scFv forms a robust interaction with an epitope consisting of two consecutive β-turns and a β-hairpin stabilized by 2 hydrogen bonds. The V H -V L interface within the 4H11-scFv is stabilized through an intricate network of 11 hydrogen bonds and a cation-π interaction. Together, our studies offer insight into antibody-MUC16 ectodomain interaction and advance our ability to design agents with potentially improved therapeutic properties over anti-CA125 moiety antibodies.
- Division of Hematology & Oncology, Department of Medicine, Massachusetts General Hospital-Harvard Medical School, Boston, MA, USA.
Organizational Affiliation: