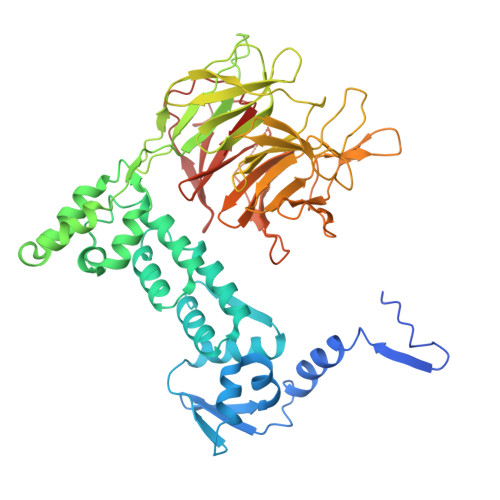
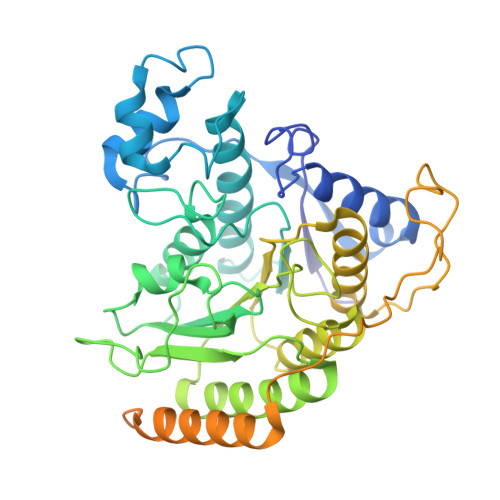
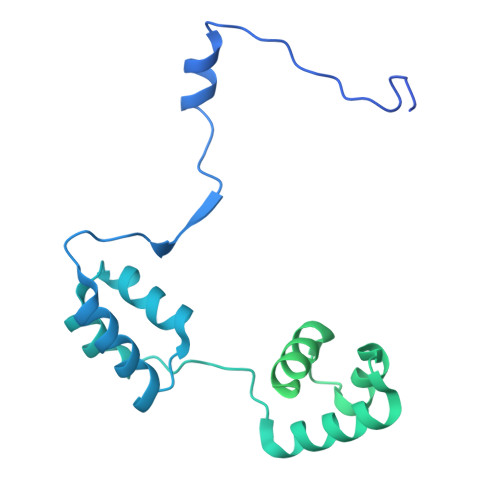
UM171 glues asymmetric CRL3-HDAC1/2 assembly to degrade CoREST corepressors.
Yeo, M.J.R., Zhang, O., Xie, X., Nam, E., Payne, N.C., Gosavi, P.M., Kwok, H.S., Iram, I., Lee, C., Li, J., Chen, N.J., Nguyen, K., Jiang, H., Wang, Z.A., Lee, K., Mao, H., Harry, S.A., Barakat, I.A., Takahashi, M., Waterbury, A.L., Barone, M., Mattevi, A., Carr, S.A., Udeshi, N.D., Bar-Peled, L., Cole, P.A., Mazitschek, R., Liau, B.B., Zheng, N.(2025) Nature 639: 232-240
- PubMed: 39939761
- DOI: https://doi.org/10.1038/s41586-024-08532-4
- Primary Citation of Related Structures:
8VOJ, 9DTG - PubMed Abstract:
UM171 is a potent agonist of ex vivo human haematopoietic stem cell self-renewal 1 . By co-opting KBTBD4, a substrate receptor of the CUL3-RING E3 ubiquitin ligase (CRL3) complex, UM171 promotes the degradation of the LSD1-CoREST corepressor complex, thereby limiting haematopoietic stem cell attrition 2,3 . However, the direct target and mechanism of action of UM171 remain unclear. Here we show that UM171 acts as a molecular glue to induce high-affinity interactions between KBTBD4 and HDAC1/2 to promote corepressor degradation. Through proteomics and chemical inhibitor studies, we identify the principal target of UM171 as HDAC1/2. Cryo-electron microscopy analysis of dimeric KBTBD4 bound to UM171 and the LSD1-HDAC1-CoREST complex identifies an asymmetric assembly in which a single UM171 molecule enables a pair of KELCH-repeat propeller domains to recruit the HDAC1 catalytic domain. One KBTBD4 propeller partially masks the rim of the HDAC1 active site, which is exploited by UM171 to extend the E3-neosubstrate interface. The other propeller cooperatively strengthens HDAC1 binding through a distinct interface. The overall CoREST-HDAC1/2-KBTBD4 interaction is further buttressed by the endogenous cofactor inositol hexakisphosphate, which acts as a second molecular glue. The functional relevance of the quaternary complex interaction surfaces is demonstrated by base editor scanning of KBTBD4 and HDAC1. By delineating the direct target of UM171 and its mechanism of action, we reveal how the cooperativity offered by a dimeric CRL3 E3 can be leveraged by a small molecule degrader.
- Department of Chemistry and Chemical Biology, Harvard University, Cambridge, MA, USA.
Organizational Affiliation: