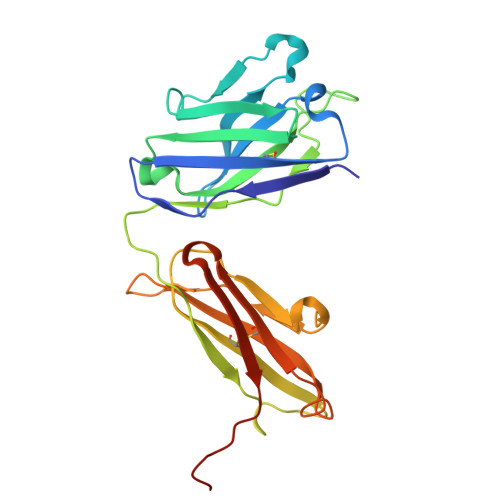
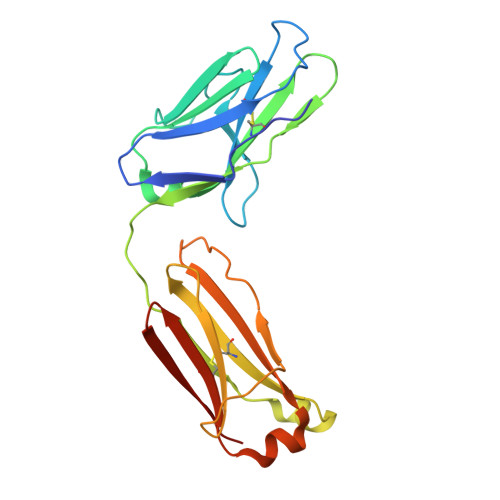
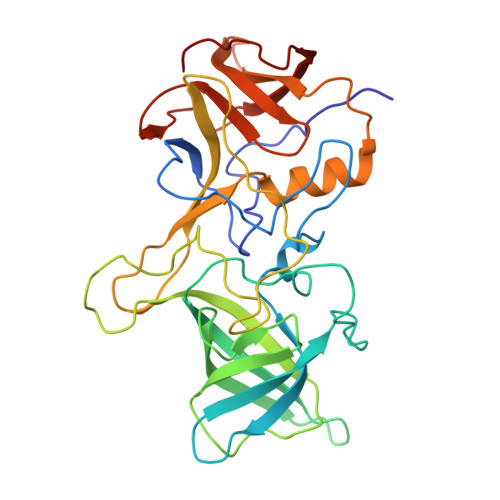
Broadly neutralizing antibodies targeting pandemic GII.4 variants or seven GII genotypes of human norovirus.
Park, J., Lindesmith, L.C., Olia, A.S., Costantini, V.P., Brewer-Jensen, P.D., Mallory, M.L., Kelley, C.E., Satterwhite, E., Longo, V., Tsybovsky, Y., Stephens, T., Marchioni, J., Martins, C.A., Huang, Y., Chaudhary, R., Zweigart, M., May, S.R., Reyes, Y., Flitter, B., Vinje, J., Tucker, S.N., Ippolito, G.C., Lavinder, J.J., Snijder, J., Kwong, P.D., Georgiou, G., Baric, R.S.(2025) Sci Transl Med 17: eads8214-eads8214
- PubMed: 40043137
- DOI: https://doi.org/10.1126/scitranslmed.ads8214
- Primary Citation of Related Structures:
8VKX - PubMed Abstract:
Human norovirus causes more than 700 million illnesses annually. Extensive genetic diversity and a paucity of information on conserved neutralizing epitopes pose major obstacles to the design of broadly protective norovirus immunogens. Here, we used high-resolution liquid chromatography-tandem mass spectrometry (LC-MS/MS)-driven proteomics to quantitatively characterize the circulating serum IgG repertoire before and after immunization with an experimental monovalent norovirus GII.4 VP1 capsid-encoding adenoviral vaccine. Two participants were specifically selected on the basis of the breadth of serum neutralization responses either across GII.4 variants (participant A) or across GII genotypes (participant B). In participant A, vaccination back-boosted highly abundant serum antibody clonotypes targeting epitopes conserved among rapidly evolving GII.4 variants spanning from a strain identified in 1987 to a strain identified in 2019. In participant B, we identified a recall response consisting of broadly neutralizing monoclonal antibodies with remarkable cross-GII ligand-binding blockade (blocking ≥ seven GII genotypes) and virus neutralization breadth. The cocrystal structure of one of these antibodies, VX22, in complex with the VP1 capsid protruding (P) domain revealed a highly conserved epitope (residues 479 to 484 and 509 to 513) within two lateral loops of the P1 subdomain. Antibody evolutionary trajectory analysis further revealed that VX22 had originally evolved from an early heterologous infection, likely by a GII.12 strain. Together, our study demonstrates that norovirus human monoclonal antibodies with broad GII.4 potency and cross-GII breadth can be boosted in serum after immunization with an adenoviral vector-based vaccine, findings that may guide the design of immunogens for broadly protective norovirus vaccines.
- Department of Molecular Biosciences, University of Texas at Austin, Austin, TX 78712, USA.
Organizational Affiliation: