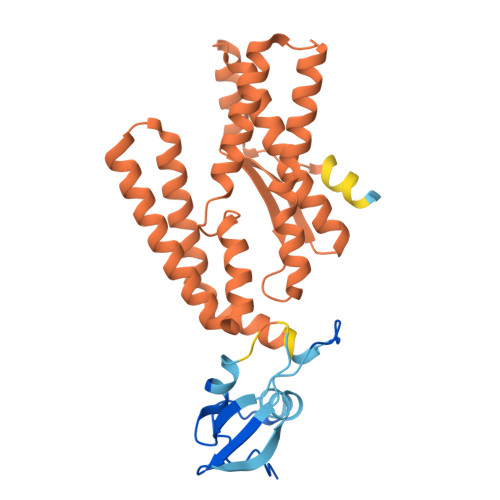
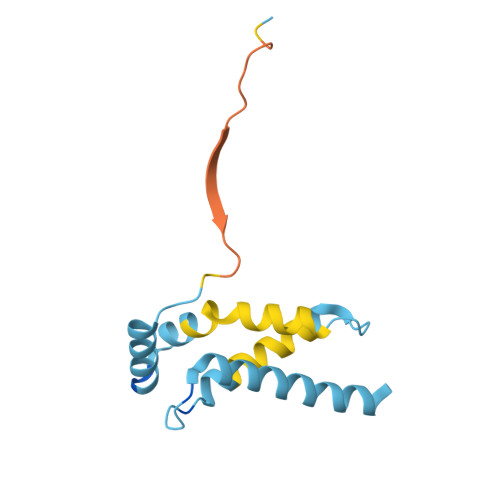
Substrate engagement by the intramembrane metalloprotease SpoIVFB.
Orlando, M.A., Pouillon, H.J.T., Mandal, S., Kroos, L., Orlando, B.J.(2024) Nat Commun 15: 8276-8276
- PubMed: 39419996
- DOI: https://doi.org/10.1038/s41467-024-52634-6
- Primary Citation of Related Structures:
8VJL, 8VJM - PubMed Abstract:
S2P intramembrane metalloproteases regulate diverse signaling pathways across all three domains of life. However, the mechanism by which S2P metalloproteases engage substrates and catalyze peptide hydrolysis within lipid membranes has remained elusive. Here we determine the cryo-EM structure of the S2P family intramembrane metalloprotease SpoIVFB from Bacillus subtilis bound to its native substrate Pro-σ K . The structure and accompanying biochemical data demonstrate that SpoIVFB positions Pro-σ K at the enzyme active site through a β-sheet augmentation mechanism, and reveal key interactions between Pro-σ K and the interdomain linker connecting SpoIVFB transmembrane and CBS domains. The cryo-EM structure and molecular dynamics simulation reveal a plausible path for water to access the membrane-buried active site of SpoIVFB, and suggest a possible role of membrane lipids in facilitating substrate capture. These results provide key insight into how S2P intramembrane metalloproteases capture and position substrates for hydrolytic proteolysis within the hydrophobic interior of a lipid membrane.
- Dept. of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI, 48824, USA.
Organizational Affiliation: