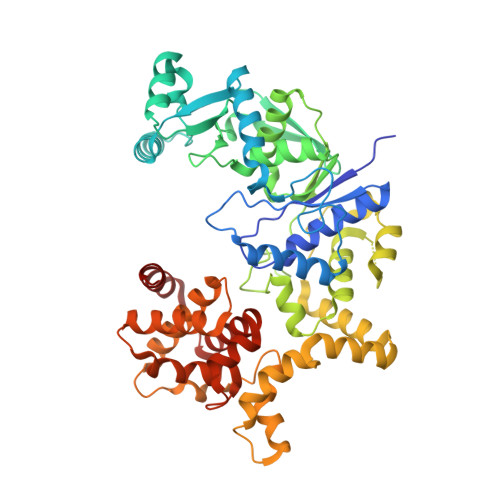
Architecture of Pseudomonas aeruginosa glutamyl-tRNA synthetase defines a subfamily of dimeric class Ib aminoacyl-tRNA synthetases.
Fenwick, M.K., Mayclin, S.J., Seibold, S., DeRocher, A.E., Subramanian, S., Phan, I.Q., Dranow, D.M., Lorimer, D.D., Abramov, A.B., Choi, R., Hewitt, S.N., Edwards, T.E., Bullard, J.M., Battaile, K.P., Wower, I.K., Soe, A.C., Tsutakawa, S.E., Lovell, S., Myler, P.J., Wower, J., Staker, B.L.(2025) Proc Natl Acad Sci U S A 122: e2504757122-e2504757122
- PubMed: 40343997
- DOI: https://doi.org/10.1073/pnas.2504757122
- Primary Citation of Related Structures:
8VC5 - PubMed Abstract:
The aminoacyl-tRNA synthetases (AaRSs) are an ancient family of structurally diverse enzymes that are divided into two major classes. The functionalities of most AaRSs are inextricably linked to their oligomeric states. While GluRSs were previously classified as monomers, the current investigation reveals that the form expressed in Pseudomonas aeruginosa is a rotationally pseudosymmetrical homodimer featuring intersubunit tRNA binding sites. Both subunits display a highly bent, "pipe strap" conformation, with the anticodon binding domain directed toward the active site. The tRNA binding sites are similar in shape to those of the monomeric GluRSs, but are formed through an approximately 180-degree rotation of the anticodon binding domains and dimerization via the anticodon and D-arm binding domains. As a result, each anticodon binding domain is poised to recognize the anticodon loop of a tRNA bound to the adjacent protomer. Additionally, the anticodon binding domain has an α-helical C -terminal extension containing a conserved lysine-rich consensus motif positioned near the predicted location of the acceptor arm, suggesting dual functions in tRNA recognition. The unique architecture of Pa GluRS broadens the structural diversity of the GluRS family, and member synthetases of all bacterial AaRS subclasses have now been identified that exhibit oligomerization.
- Seattle Structural Genomics Center for Infectious Disease, Seattle Children's Research Institute, Seattle, WA 98109.
Organizational Affiliation: