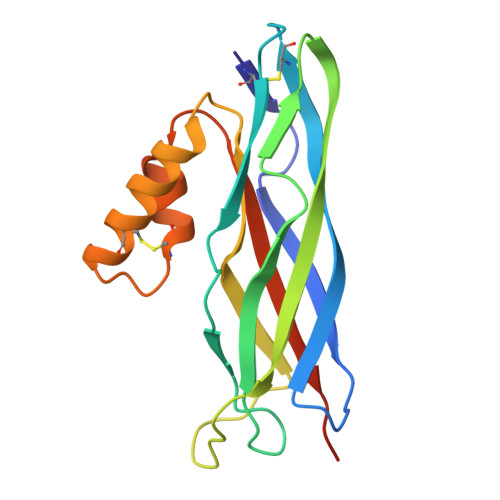
The Yeh pilus adhesin is equipped with an alpha-helical flap motif, which contributes to pectin adherence.
Tamadonfar, K.O., Klein, R.D., Lopatto, E.D.B., Zimmerman, M.I., Azimzadeh, P.N., Sanick, D.A., Porter, J.R., Villicana, J.B., Pinkner, J.S., Chiang, B.H., Gualberto, N.C., Dodson, K.W., Patnode, M.L., Birchenough, G.M.H., Bowman, G.R., Hultgren, S.J.(2026) Sci Adv 12: eadz1301-eadz1301
- PubMed: 41499512
- DOI: https://doi.org/10.1126/sciadv.adz1301
- Primary Citation of Related Structures:
8V9V, 9N4G, 9N4H, 9N4I - PubMed Abstract:
The Yeh chaperone-usher pathway (CUP) pilus adhesin is encoded in one-half of all Escherichia coli . Yet little is known about its structure and function in E. coli persistence and pathogenesis. Structural investigations reveal that the adhesin receptor binding domains (RBDs) of YehD and its relative YhlD both share a canonical β-rich core and an α-helical flap motif that is hinged at the distal end of the core. This flap was observed in both open and closed conformations using molecular dynamics simulations. The closed conformation is dependent on a hydrophobic patch of amino acids on the distal end of the flap. Functionally, YehD RBD is able to bind pectin, a polysaccharide ubiquitous in plant material. Mutations that interrupt the closed conformation increase the affinity of the protein to pectin, suggesting that the flap contributes mechanistically to pectin binding. Furthermore, in vivo, the pilus contributes to gastrointestinal (GI) tract colonization in the absence of the type 1 pilus. Hence, we report the ability of YehD to bind pectin representing a possible colonization mechanism of the GI tract via a structurally distinct CUP adhesin.
- Department of Molecular Microbiology and Center for Women's Infectious Disease Research, Washington University School of Medicine, St. Louis, MO, USA.
Organizational Affiliation: