Yerba mate ( Ilex paraguariensis ) genome provides new insights into convergent evolution of caffeine biosynthesis.
Vignale, F.A., Hernandez Garcia, A., Modenutti, C.P., Sosa, E.J., Defelipe, L.A., Oliveira, R., Nunes, G.L., Acevedo, R.M., Burguener, G.F., Rossi, S.M., Zapata, P.D., Marti, D.A., Sansberro, P., Oliveira, G., Catania, E.M., Smith, M.N., Dubs, N.M., Nair, S., Barkman, T.J., Turjanski, A.G.(2025) Elife 14
- PubMed: 39773819
- DOI: https://doi.org/10.7554/eLife.104759
- Primary Citation of Related Structures:
8UZD - PubMed Abstract:
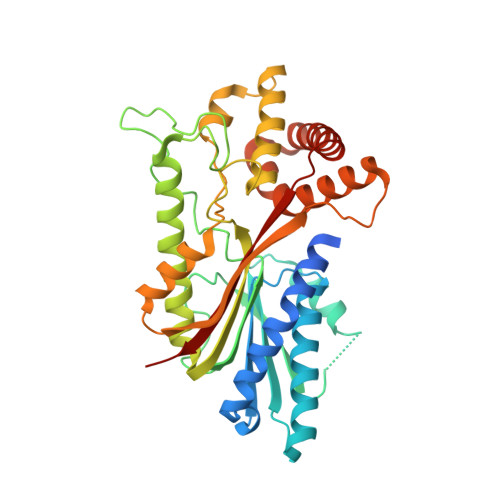
Yerba mate (YM, Ilex paraguariensis ) is an economically important crop marketed for the elaboration of mate, the third-most widely consumed caffeine-containing infusion worldwide. Here, we report the first genome assembly of this species, which has a total length of 1.06 Gb and contains 53,390 protein-coding genes. Comparative analyses revealed that the large YM genome size is partly due to a whole-genome duplication (Ip-α) during the early evolutionary history of Ilex , in addition to the hexaploidization event (γ) shared by core eudicots. Characterization of the genome allowed us to clone the genes encoding methyltransferase enzymes that catalyse multiple reactions required for caffeine production. To our surprise, this species has converged upon a different biochemical pathway compared to that of coffee and tea. In order to gain insight into the structural basis for the convergent enzyme activities, we obtained a crystal structure for the terminal enzyme in the pathway that forms caffeine. The structure reveals that convergent solutions have evolved for substrate positioning because different amino acid residues facilitate a different substrate orientation such that efficient methylation occurs in the independently evolved enzymes in YM and coffee. While our results show phylogenomic constraint limits the genes coopted for convergence of caffeine biosynthesis, the X-ray diffraction data suggest structural constraints are minimal for the convergent evolution of individual reactions.
- European Molecular Biology Laboratory - Hamburg Unit, Hamburg, Germany.
Organizational Affiliation: