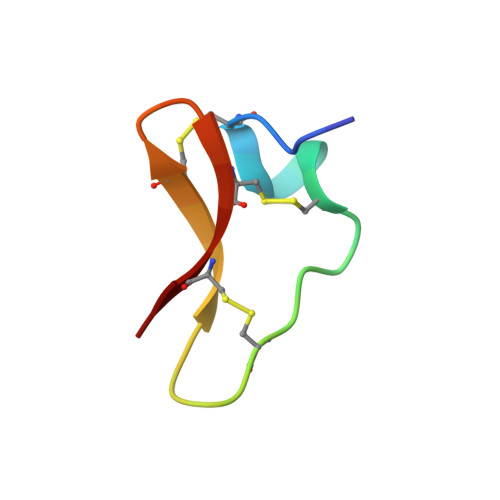
Chickpea NCR13 disulfide cross-linking variants exhibit profound differences in antifungal activity and modes of action.
Godwin, J., Djami-Tchatchou, A.T., Velivelli, S.L.S., Tetorya, M., Kalunke, R., Pokhrel, A., Zhou, M., Buchko, G.W., Czymmek, K.J., Shah, D.M.(2024) PLoS Pathog 20: e1012745-e1012745
- PubMed: 39621770
- DOI: https://doi.org/10.1371/journal.ppat.1012745
- Primary Citation of Related Structures:
7TH8, 8ULM - PubMed Abstract:
Small cysteine-rich antifungal peptides with multi-site modes of action (MoA) have potential for development as biofungicides. In particular, legumes of the inverted repeat-lacking clade express a large family of nodule-specific cysteine-rich (NCR) peptides that orchestrate differentiation of nitrogen-fixing bacteria into bacteroids. These NCRs can form two or three intramolecular disulfide bonds and a subset of these peptides with high cationicity exhibits antifungal activity. However, the importance of intramolecular disulfide pairing and MoA against fungal pathogens for most of these plant peptides remains to be elucidated. Our study focused on a highly cationic chickpea NCR13, which has a net charge of +8 and contains six cysteines capable of forming three disulfide bonds. NCR13 expression in Pichia pastoris resulted in formation of two peptide folding variants, NCR13_PFV1 and NCR13_PFV2, that differed in the pairing of two out of three disulfide bonds despite having an identical amino acid sequence. The NMR structure of each PFV revealed a unique three-dimensional fold with the PFV1 structure being more compact but less dynamic. Surprisingly, PFV1 and PFV2 differed profoundly in the potency of antifungal activity against several fungal plant pathogens and their multi-faceted MoA. PFV1 showed significantly faster fungal cell-permeabilizing and cell entry capabilities as well as greater stability once inside the fungal cells. Additionally, PFV1 was more effective in binding fungal ribosomal RNA and inhibiting protein translation in vitro. Furthermore, when sprayed on pepper and tomato plants, PFV1 was more effective in reducing disease symptoms caused by Botrytis cinerea, causal agent of gray mold disease in fruits, vegetables, and flowers. In conclusion, our work highlights the significant impact of disulfide pairing on the antifungal activity and MoA of NCR13 and provides a structural framework for design of novel, potent antifungal peptides for agricultural use.
- Donald Danforth Plant Science Center, St. Louis, Missouri, United States of America.
Organizational Affiliation: