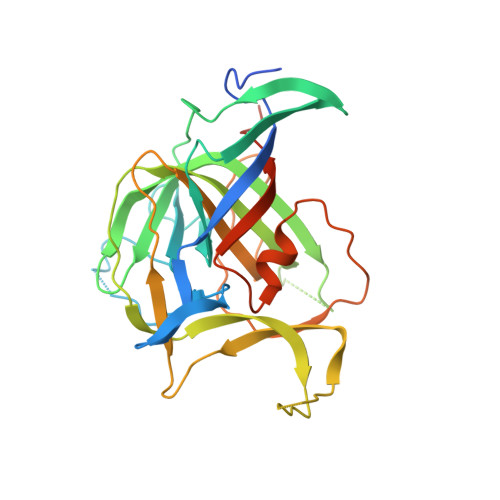
Structure and antigenicity of the divergent human astrovirus VA1 capsid spike.
Ghosh, A., Delgado-Cunningham, K., Lopez, T., Green, K., Arias, C.F., DuBois, R.M.(2024) PLoS Pathog 20: e1012028-e1012028
- PubMed: 38416796
- DOI: https://doi.org/10.1371/journal.ppat.1012028
- Primary Citation of Related Structures:
8UFN, 8UFO - PubMed Abstract:
Human astrovirus (HAstV) is a known cause of viral gastroenteritis in children worldwide, but HAstV can cause also severe and systemic infections in immunocompromised patients. There are three clades of HAstV: classical, MLB, and VA/HMO. While all three clades are found in gastrointestinal samples, HAstV-VA/HMO is the main clade associated with meningitis and encephalitis in immunocompromised patients. To understand how the HAstV-VA/HMO can infect the central nervous system, we investigated its sequence-divergent capsid spike, which functions in cell attachment and may influence viral tropism. Here we report the high-resolution crystal structures of the HAstV-VA1 capsid spike from strains isolated from patients with gastrointestinal and neuronal disease. The HAstV-VA1 spike forms a dimer and shares a core beta-barrel structure with other astrovirus capsid spikes but is otherwise strikingly different, suggesting that HAstV-VA1 may utilize a different cell receptor, and an infection competition assay supports this hypothesis. Furthermore, by mapping the capsid protease cleavage site onto the structure, the maturation and assembly of the HAstV-VA1 capsid is revealed. Finally, comparison of gastrointestinal and neuronal HAstV-VA1 sequences, structures, and antigenicity suggests that neuronal HAstV-VA1 strains may have acquired immune escape mutations. Overall, our studies on the HAstV-VA1 capsid spike lay a foundation to further investigate the biology of HAstV-VA/HMO and to develop vaccines and therapeutics targeting it.
- Department of Biomolecular Engineering, University of California Santa Cruz, Santa Cruz, California, United States of America.
Organizational Affiliation: