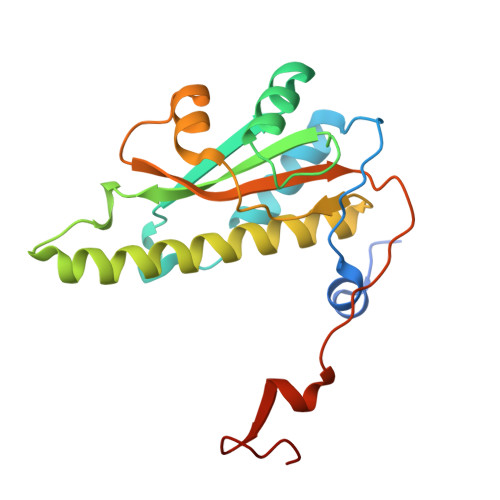
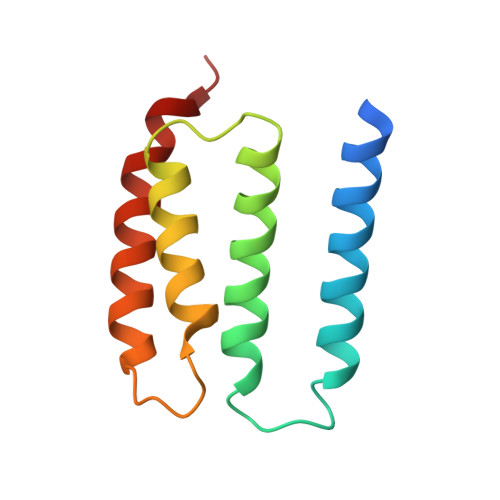
Cyclodipeptide oxidase is an enzyme filament.
Andreas, M.P., Giessen, T.W.(2024) Nat Commun 15: 3574-3574
- PubMed: 38678027
- DOI: https://doi.org/10.1038/s41467-024-48030-9
- Primary Citation of Related Structures:
8UC3 - PubMed Abstract:
Modified cyclic dipeptides represent a widespread class of secondary metabolites with diverse pharmacological activities, including antibacterial, antifungal, and antitumor. Here, we report the structural characterization of the Streptomyces noursei enzyme AlbAB, a cyclodipeptide oxidase (CDO) carrying out α,β-dehydrogenations during the biosynthesis of the antibiotic albonoursin. We show that AlbAB is a megadalton heterooligomeric enzyme filament containing covalently bound flavin mononucleotide cofactors. We highlight that AlbAB filaments consist of alternating dimers of AlbA and AlbB and that enzyme activity is crucially dependent on filament formation. We show that AlbA-AlbB interactions are highly conserved suggesting that other CDO-like enzymes are likely enzyme filaments. As CDOs have been employed in the structural diversification of cyclic dipeptides, our results will be useful for future applications of CDOs in biocatalysis and chemoenzymatic synthesis.
- Department of Biological Chemistry, University of Michigan Medical School, Ann Arbor, MI, 48109, USA.
Organizational Affiliation: