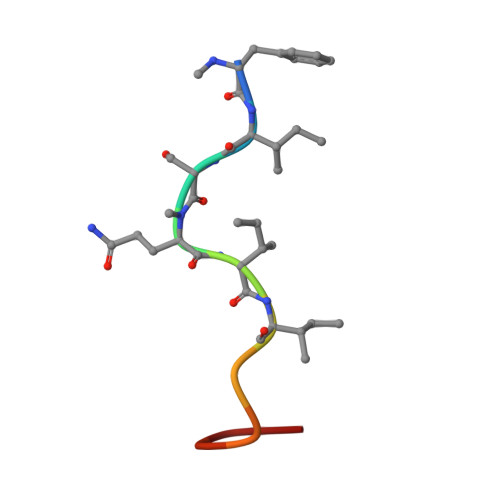
Supramolecular Interactions of Teixobactin Analogues in the Crystal State.
Yang, H., Kreutzer, A.G., Nowick, J.S.(2024) J Org Chem 89: 5104-5108
- PubMed: 38506062
- DOI: https://doi.org/10.1021/acs.joc.3c02617
- Primary Citation of Related Structures:
8U78 - PubMed Abstract:
This Note presents the X-ray crystallographic structure of the N -methylated teixobactin analogue N -Me-d-Gln 4 ,Lys 10 -teixobactin ( 1 ). Eight peptide molecules comprise the asymmetric unit, with each peptide molecule binding a chloride anion through hydrogen bonding with the amide NH group of residues 7, 8, 10, and 11. The peptide molecules form hydrogen-bonded antiparallel β-sheet dimers in the crystal lattice, with residues 1-3 comprising the dimerization interface. The dimers further assemble end-to-end in the crystal lattice.
- Department of Chemistry, University of California Irvine, Irvine, California 92697, United States.
Organizational Affiliation: