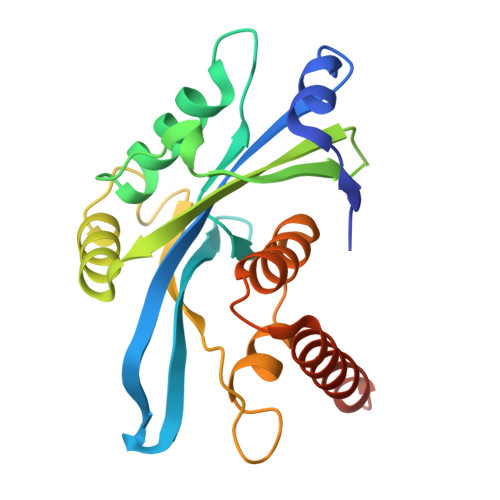
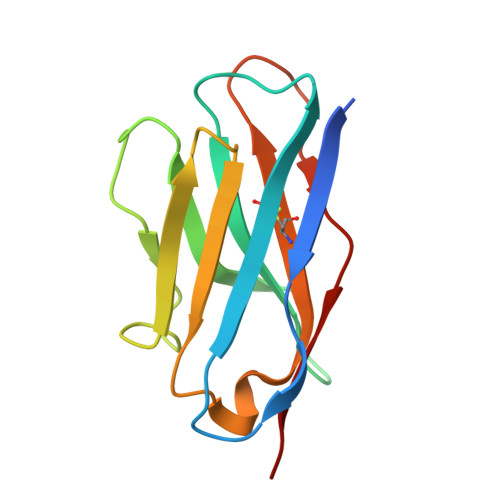
NEAT1 modulates the TIRR/53BP1 complex to maintain genome integrity.
Kilgas, S., Syed, A., Toolan-Kerr, P., Swift, M.L., Roychoudhury, S., Sarkar, A., Wilkins, S., Quigley, M., Poetsch, A.R., Botuyan, M.V., Cui, G., Mer, G., Ule, J., Drane, P., Chowdhury, D.(2024) Nat Commun 15: 8438-8438
- PubMed: 39349456
- DOI: https://doi.org/10.1038/s41467-024-52862-w
- Primary Citation of Related Structures:
8U3S - PubMed Abstract:
Tudor Interacting Repair Regulator (TIRR) is an RNA-binding protein (RBP) that interacts directly with 53BP1, restricting its access to DNA double-strand breaks (DSBs) and its association with p53. We utilized iCLIP to identify RNAs that directly bind to TIRR within cells, identifying the long non-coding RNA NEAT1 as the primary RNA partner. The high affinity of TIRR for NEAT1 is due to prevalent G-rich motifs in the short isoform (NEAT1_1) region of NEAT1. This interaction destabilizes the TIRR/53BP1 complex, promoting 53BP1's function. NEAT1_1 is enriched during the G1 phase of the cell cycle, thereby ensuring that TIRR-dependent inhibition of 53BP1's function is cell cycle-dependent. TDP-43, an RBP that is implicated in neurodegenerative diseases, modulates the TIRR/53BP1 complex by promoting the production of the NEAT1 short isoform, NEAT1_1. Together, we infer that NEAT1_1, and factors regulating NEAT1_1, may impact 53BP1-dependent DNA repair processes, with implications for a spectrum of diseases.
- Division of Radiation and Genome Stability, Department of Radiation Oncology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA, USA.
Organizational Affiliation: