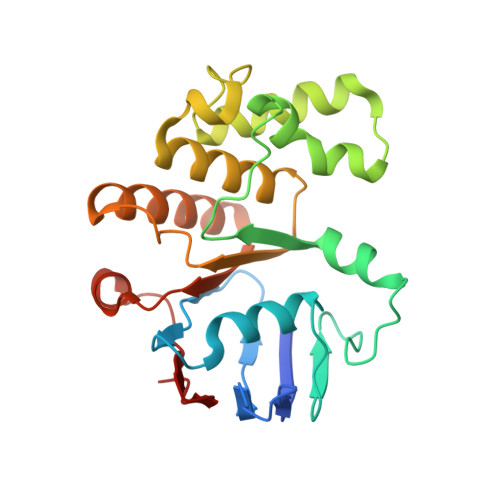
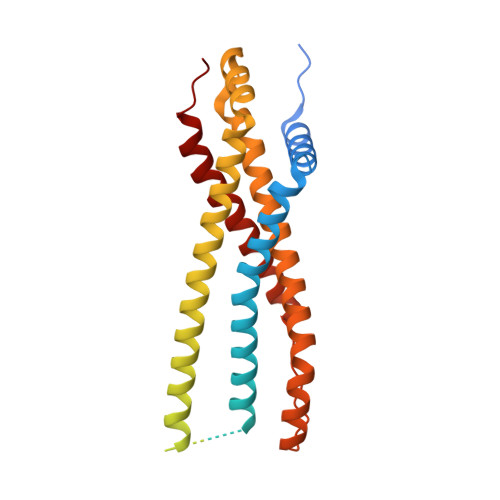
Structural insights into the FtsEX-EnvC complex regulation on septal peptidoglycan hydrolysis in Vibrio cholerae.
Hao, A., Suo, Y., Lee, S.Y.(2024) Structure 32: 188-199.e5
- PubMed: 38070498
- DOI: https://doi.org/10.1016/j.str.2023.11.007
- Primary Citation of Related Structures:
8TZJ, 8TZK, 8TZL - PubMed Abstract:
During bacterial cell division, hydrolysis of septal peptidoglycan (sPG) is crucial for cell separation. This sPG hydrolysis is performed by the enzyme amidases whose activity is regulated by the integral membrane protein complex FtsEX-EnvC. FtsEX is an ATP-binding cassette transporter, and EnvC is a long coiled-coil protein that interacts with and activates the amidases. The molecular mechanism by which the FtsEX-EnvC complex activates amidases remains largely unclear. We present the cryo-electron microscopy structure of the FtsEX-EnvC complex from the pathogenic bacteria V. cholerae (FtsEX-EnvC VC ). FtsEX-EnvC VC in the presence of ADP adopts a distinct conformation where EnvC is "horizontally extended" rather than "vertically extended". Subsequent structural studies suggest that EnvC can swing between these conformations in space in a nucleotide-dependent manner. Our structural analysis and functional studies suggest that FtsEX-EnvC VC employs spatial control of EnvC for amidase activation, providing mechanistic insights into the FtsEX-EnvC regulation on septal peptidoglycan hydrolysis.
- Department of Biochemistry, Duke University Medical Center, Durham, NC 27710, USA.
Organizational Affiliation: