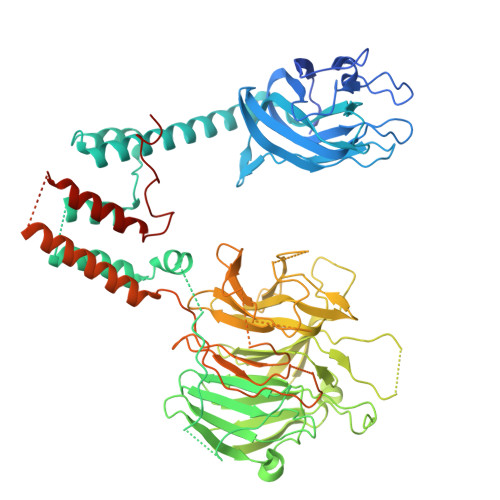
FAM72A degrades UNG2 through the GID/CTLH complex to promote mutagenic repair during antibody maturation.
Barbulescu, P., Chana, C.K., Wong, M.K., Ben Makhlouf, I., Bruce, J.P., Feng, Y., Keszei, A.F.A., Wong, C., Mohamad-Ramshan, R., McGary, L.C., Kashem, M.A., Ceccarelli, D.F., Orlicky, S., Fang, Y., Kuang, H., Mazhab-Jafari, M., Pezo, R.C., Bhagwat, A.S., Pugh, T.J., Gingras, A.C., Sicheri, F., Martin, A.(2024) Nat Commun 15: 7541-7541
- PubMed: 39215025
- DOI: https://doi.org/10.1038/s41467-024-52009-x
- Primary Citation of Related Structures:
8TTQ - PubMed Abstract:
A diverse antibody repertoire is essential for humoral immunity. Antibody diversification requires the introduction of deoxyuridine (dU) mutations within immunoglobulin genes to initiate somatic hypermutation (SHM) and class switch recombination (CSR). dUs are normally recognized and excised by the base excision repair (BER) protein uracil-DNA glycosylase 2 (UNG2). However, FAM72A downregulates UNG2 permitting dUs to persist and trigger SHM and CSR. How FAM72A promotes UNG2 degradation is unknown. Here, we show that FAM72A recruits a C-terminal to LisH (CTLH) E3 ligase complex to target UNG2 for proteasomal degradation. Deficiency in CTLH complex components result in elevated UNG2 and reduced SHM and CSR. Cryo-EM structural analysis reveals FAM72A directly binds to MKLN1 within the CTLH complex to recruit and ubiquitinate UNG2. Our study further suggests that FAM72A hijacks the CTLH complex to promote mutagenesis in cancer. These findings show that FAM72A is an E3 ligase substrate adaptor critical for humoral immunity and cancer development.
- Department of Immunology, University of Toronto, Toronto, ON, Canada.
Organizational Affiliation: