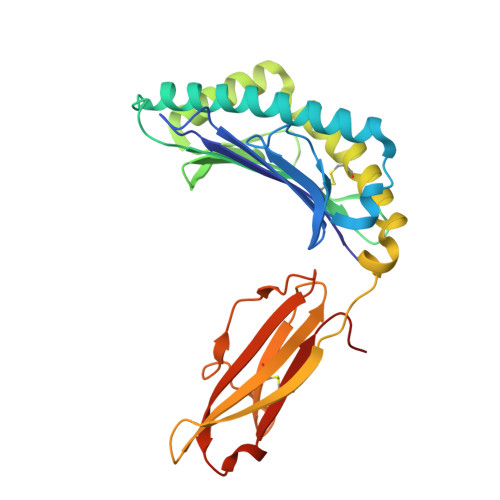
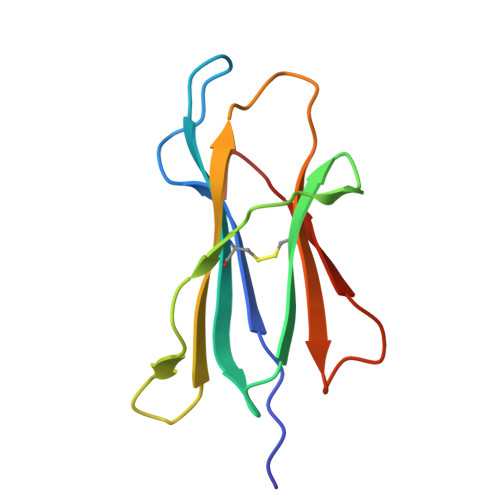
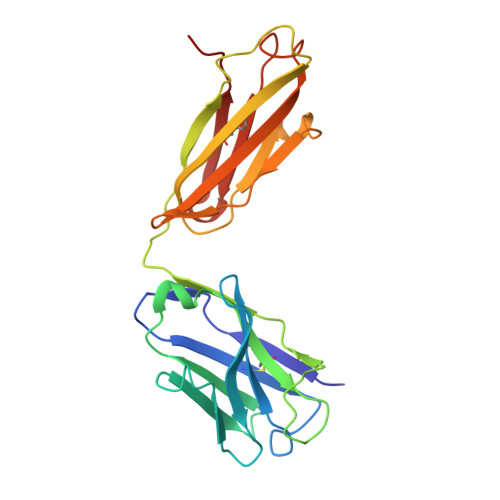
Antibody Mediated Inhibition of HLA/LILR Interactions Breaks Innate Immune Tolerance and Induces Antitumor Immunity.
Panda, A.K., Natarajan, K., Sinha, S., Jiang, J., Chempati, S., Boyd, L.F., Desai, P.P., Buszko, M., Kim, Y.H., Kazmi, S., Fisk, B., Teke, M.E., Larrain, C.M., Remmert, K., Blakely, A.M., Douagi, I., Hernandez, J.M., Margulies, D.H., Shevach, E.M.(2025) Cancer Immunol Res
- PubMed: 41032026
- DOI: https://doi.org/10.1158/2326-6066.CIR-25-0343
- Primary Citation of Related Structures:
8TQ5 - PubMed Abstract:
Immune check-point blockade for the treatment of malignancies has been focused on reversing inhibitory pathways in T lymphocytes. Natural killer (NK) cells are a potent innate defense against tumors and virally infected cells, but their therapeutic manipulation for anti-cancer immunity has been inadequately explored. Considerable attention has been focused on approaches to blocking inhibitory receptors on NK and myeloid cells. Most effort has been directed to the killer immunoglobulin-like receptors (KIR) and CD94/NKG2A on NK cells. Another set of receptors with similar function in both NK cells and myeloid cells is the leukocyte immunoglobulin like receptors (LILR) that interact with a wide variety of HLA molecules. Using pan-anti-HLA mAbs that recognize a conserved epitopic region on HLA also seen by LILR, we investigated their functional effects in several models of tumor immunity. The pan-anti-HLA-mAbs blocked the binding of most LILRs, did not block killer cell immunoglobulin-like receptors (KIR) or CD94/NKG2A/C or TCR recognition. They also activated dysfunctional NK cells explanted from a variety of human cancers, and resulted in enhancement of tumor immunity in humanized mice. The mAbs also exert direct anti-tumor effects. These results suggest that activation of innate immunity via disruption of HLA/LILR interactions is a potent approach for control of both primary tumors and potentially tumor metastases.
- National Institute of Allergy and Infectious Diseases, Bethesda, Maryland, United States.
Organizational Affiliation: