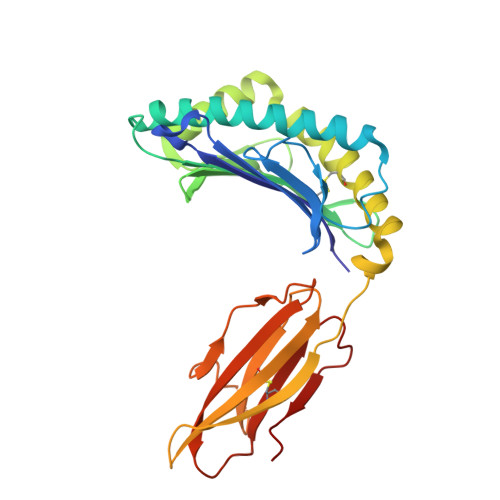
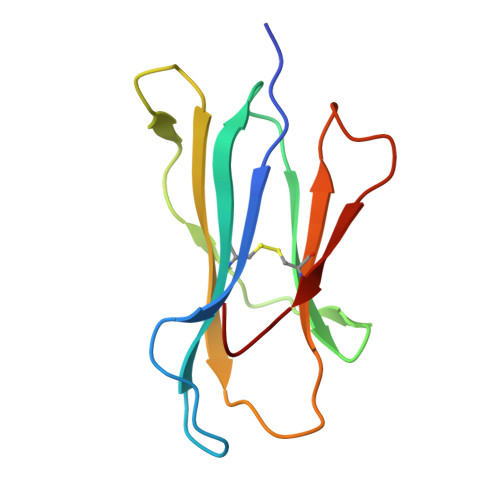
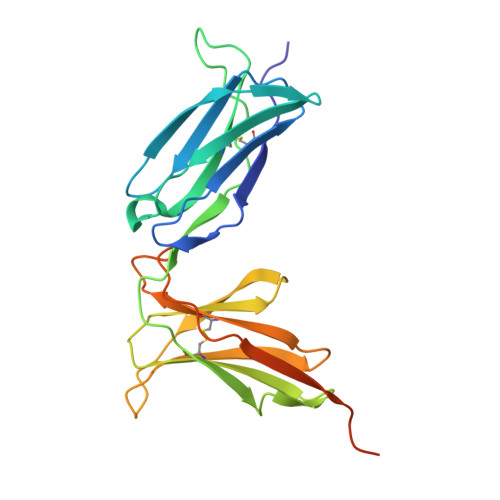
Molecular characterization of the archaic HLA-B∗73:01 allele reveals presentation of a unique peptidome and skewed engagement by KIR2DL2.
Ross, P., Hilton, H.G., Lodwick, J., Slezak, T., Guethlein, L.A., McMurtrey, C.P., Han, A.S., Nielsen, M., Yong, D., Dulberger, C.L., Nolan, K.T., Roy, S., Castro, C.D., Hildebrand, W.H., Zhao, M., Kossiakoff, A., Parham, P., Adams, E.J.(2025) J Biological Chem 301: 110542-110542
- PubMed: 40749828
- DOI: https://doi.org/10.1016/j.jbc.2025.110542
- Primary Citation of Related Structures:
8TMU - PubMed Abstract:
HLA class I alleles of archaic origin may have been retained in modern humans because they provide immunity against diseases to which archaic humans had evolved resistance. According to this model, archaic introgressed alleles were somehow distinct from those that evolved in African populations. Here, we show that HLA-B∗73:01, a rare allotype with putative archaic origins, has a relatively rare peptide binding motif with an unusually long-tailed peptide length distribution. We also find that HLA-B∗73:01 combines a restricted and unique peptidome with high-cell surface expression, characteristics that make it well-suited to combat one or a number of closely related pathogens. Furthermore, a crystal structure of HLA-B∗73:01 in complex with KIR2DL2 highlights differences from previously solved structures with HLA-C molecules. These molecular characteristics distinguish HLA-B∗73:01 from other HLA class I alleles previously investigated and may have provided early modern human migrants that inherited this allele with a selective advantage as they colonized Europe and Asia.
- Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, Illinois, USA; Committee on Genetics, Genomics and Systems Biology, The University of Chicago, Chicago, Illinois, USA.
Organizational Affiliation: