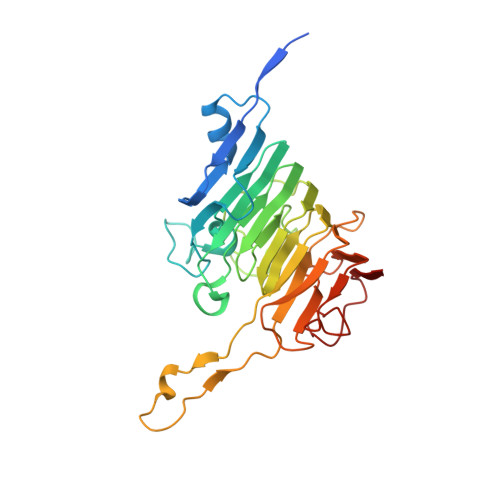
Crystal Structures of Bacterial Pectin Methylesterases Pme8A and PmeC2 from Rumen Butyrivibrio .
Carbone, V., Reilly, K., Sang, C., Schofield, L.R., Ronimus, R.S., Kelly, W.J., Attwood, G.T., Palevich, N.(2023) Int J Mol Sci 24
- PubMed: 37762041
- DOI: https://doi.org/10.3390/ijms241813738
- Primary Citation of Related Structures:
8TMS, 8TNE - PubMed Abstract:
Pectin is a complex polysaccharide that forms a substantial proportion of the plant's middle lamella of forage ingested by grazing ruminants. Methanol in the rumen is derived mainly from methoxy groups released from pectin by the action of pectin methylesterase (PME) and is subsequently used by rumen methylotrophic methanogens that reduce methanol to produce methane (CH 4 ). Members of the genus Butyrivibrio are key pectin-degrading rumen bacteria that contribute to methanol formation and have important roles in fibre breakdown, protein digestion, and the biohydrogenation of fatty acids. Therefore, methanol release from pectin degradation in the rumen is a potential target for CH 4 mitigation technologies. Here, we present the crystal structures of PMEs belonging to the carbohydrate esterase family 8 (CE8) from Butyrivibrio proteoclasticus and Butyrivibrio fibrisolvens , determined to a resolution of 2.30 Å. These enzymes, like other PMEs, are right-handed β-helical proteins with a well-defined catalytic site and reaction mechanisms previously defined in insect, plant, and other bacterial pectin methylesterases. Potential substrate binding domains are also defined for the enzymes.
- AgResearch Limited, Grasslands Research Centre, Palmerston North 4442, New Zealand.
Organizational Affiliation: