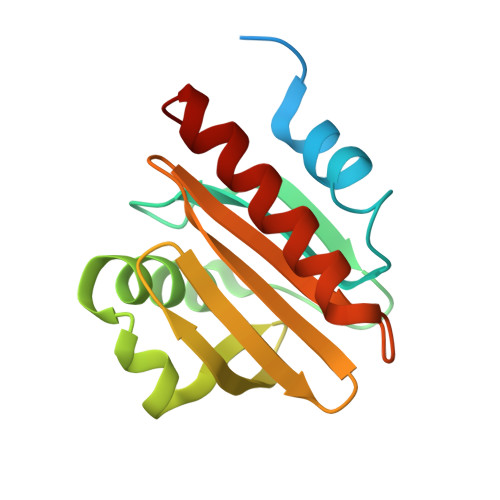
Structural homology of mite profilins to plant profilins is not indicative of allergic cross-reactivity.
O'Malley, A., Sankaran, S., Carriuolo, A., Khatri, K., Kowal, K., Chruszcz, M.(2024) Biol Chem 405: 367-381
- PubMed: 38662449
- DOI: https://doi.org/10.1515/hsz-2023-0366
- Primary Citation of Related Structures:
8TI5, 8TI6, 8TI7, 8V5Y - PubMed Abstract:
Structural and allergenic characterization of mite profilins has not been previously pursued to a similar extent as plant profilins. Here, we describe structures of profilins originating from Tyrophagus putrescentiae (registered allergen Tyr p 36.0101) and Dermatophagoides pteronyssinus (here termed Der p profilin), which are the first structures of profilins from Arachnida. Additionally, the thermal stabilities of mite and plant profilins are compared, suggesting that the high number of cysteine residues in mite profilins may play a role in their increased stability. We also examine the cross-reactivity of plant and mite profilins as well as investigate the relevance of these profilins in mite inhalant allergy. Despite their high structural similarity to other profilins, mite profilins have low sequence identity with plant and human profilins. Subsequently, these mite profilins most likely do not display cross-reactivity with plant profilins. At the same time the profilins have highly conserved poly(l-proline) and actin binding sites.
- Department of Biochemistry and Molecular Biology, 3078 Michigan State University , 603 Wilson Road, East Lansing, MI 48824, USA.
Organizational Affiliation: