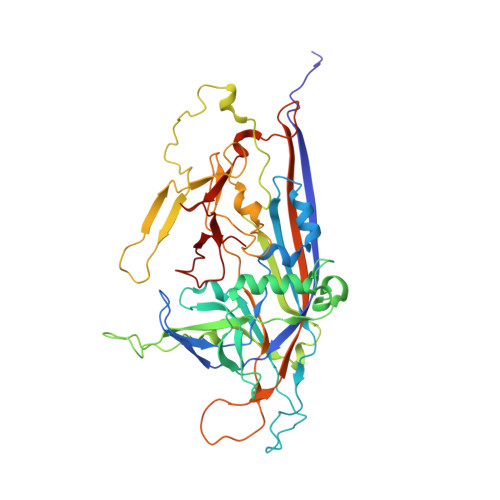
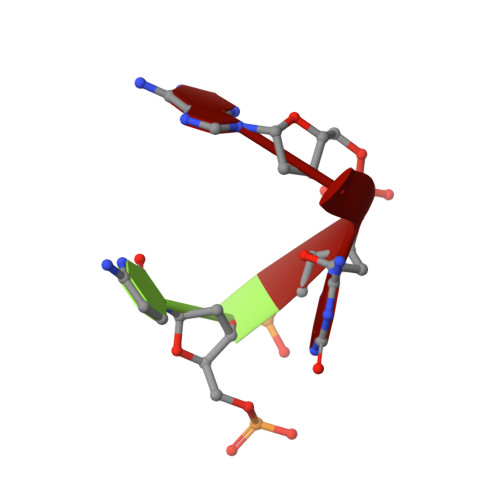
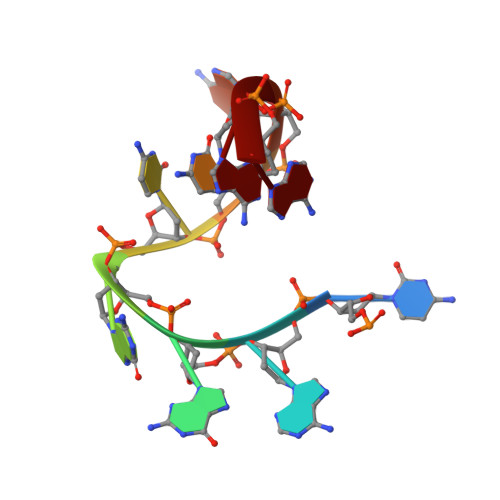
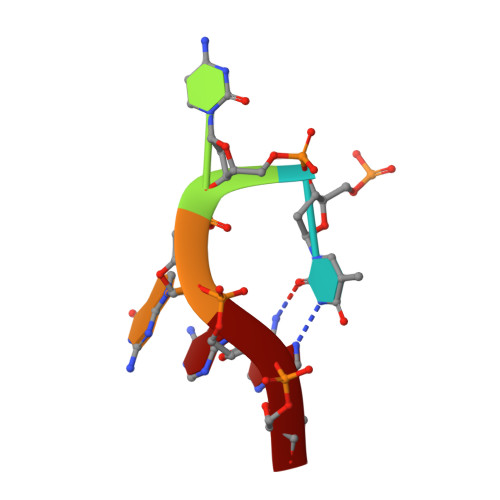
Cryo-EM-based discovery of a pathogenic parvovirus causing epidemic mortality by black wasting disease in farmed beetles.
Penzes, J.J., Holm, M., Yost, S.A., Kaelber, J.T.(2024) Cell 187: 5604-5619.e14
- PubMed: 39208798
- DOI: https://doi.org/10.1016/j.cell.2024.07.053
- Primary Citation of Related Structures:
8T9C, 8T9E, 8T9X, 8TA7, 8TJE - PubMed Abstract:
We use cryoelectron microscopy (cryo-EM) as a sequence- and culture-independent diagnostic tool to identify the etiological agent of an agricultural pandemic. For the past 4 years, American insect-rearing facilities have experienced a distinctive larval pathology and colony collapse of farmed Zophobas morio (superworm). By means of cryo-EM, we discovered the causative agent: a densovirus that we named Zophobas morio black wasting virus (ZmBWV). We confirmed the etiology of disease by fulfilling Koch's postulates and characterizing strains from across the United States. ZmBWV is a member of the family Parvoviridae with a 5,542 nt genome, and we describe intersubunit interactions explaining its expanded internal volume relative to human parvoviruses. Cryo-EM structures at resolutions up to 2.1 Å revealed single-strand DNA (ssDNA) ordering at the capsid inner surface pinned by base-binding pockets in the capsid inner surface. Also, we demonstrated the prophylactic potential of non-pathogenic strains to provide cross-protection in vivo.
- Institute for Quantitative Biomedicine, Rutgers, The State University of New Jersey, Piscataway, NJ, USA. Electronic address: judit.penzes@rutgers.edu.
Organizational Affiliation: