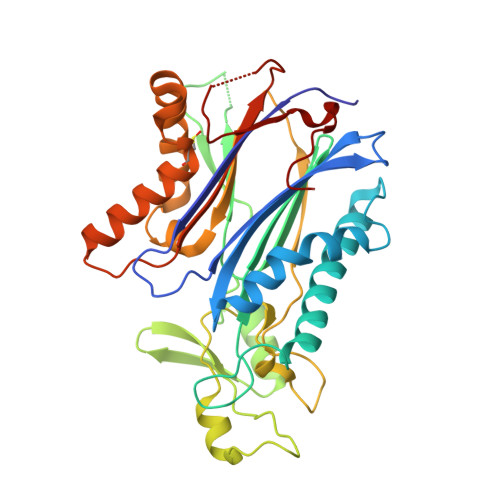
Crystal structure and mechanistic studies of the PPM1D serine/threonine phosphatase catalytic domain.
Kumar, J.P., Kosek, D., Durell, S.R., Miller Jenkins, L.M., Debnath, S., Coussens, N.P., Hall, M.D., Appella, D.H., Dyda, F., Mazur, S.J., Appella, E.(2024) J Biological Chem 300: 107561-107561
- PubMed: 39002674
- DOI: https://doi.org/10.1016/j.jbc.2024.107561
- Primary Citation of Related Structures:
8T2J - PubMed Abstract:
Protein phosphatase 1D (PPM1D, Wip1) is induced by the tumor suppressor p53 during DNA damage response signaling and acts as an oncoprotein in several human cancers. Although PPM1D is a potential therapeutic target, insights into its atomic structure were challenging due to flexible regions unique to this family member. Here we report the first crystal structure of the PPM1D catalytic domain to 1.8 Å resolution. The structure reveals the active site with two Mg 2+ ions bound, similar to other structures. The flap subdomain and B-loop, which are crucial for substrate recognition and catalysis, were also resolved, with the flap forming two short helices and three short β-strands that are followed by an irregular loop. Unexpectedly, a nitrogen-oxygen-sulfur bridge was identified in the catalytic domain. Molecular dynamics simulations and kinetic studies provided further mechanistic insights into the regulation of PPM1D catalytic activity. In particular, the kinetic experiments demonstrated a magnesium concentration-dependent lag in PPM1D attaining steady-state velocity, a feature of hysteretic enzymes that show slow transitions compared with catalytic turnover. All combined, these results advance the understanding of PPM1D function and will support the development of PPM1D-targeted therapeutics.
- Laboratory of Cell Biology, NCI, National Institutes of Health, Bethesda, MD 20892, United States.
Organizational Affiliation: