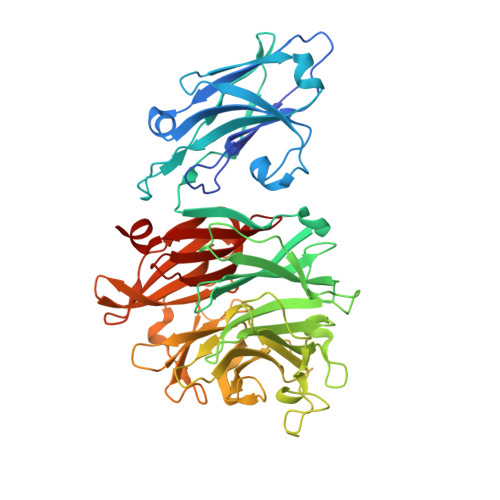
Functional and structural analyses reveal that a dual domain sialidase protects bacteria from complement killing through desialylation of complement factors.
Clark, N.D., Pham, C., Kurniyati, K., Sze, C.W., Coleman, L., Fu, Q., Zhang, S., Malkowski, M.G., Li, C.(2023) PLoS Pathog 19: e1011674-e1011674
- PubMed: 37747935
- DOI: https://doi.org/10.1371/journal.ppat.1011674
- Primary Citation of Related Structures:
8FEB, 8T1Y, 8T1Z, 8T24, 8T26, 8T27 - PubMed Abstract:
The complement system is the first line of innate immune defense against microbial infections. To survive in humans and cause infections, bacterial pathogens have developed sophisticated mechanisms to subvert the complement-mediated bactericidal activity. There are reports that sialidases, also known as neuraminidases, are implicated in bacterial complement resistance; however, its underlying molecular mechanism remains elusive. Several complement proteins (e.g., C1q, C4, and C5) and regulators (e.g., factor H and C4bp) are modified by various sialoglycans (glycans with terminal sialic acids), which are essential for their functions. This report provides both functional and structural evidence that bacterial sialidases can disarm the complement system via desialylating key complement proteins and regulators. The oral bacterium Porphyromonas gingivalis, a "keystone" pathogen of periodontitis, produces a dual domain sialidase (PG0352). Biochemical analyses reveal that PG0352 can desialylate human serum and complement factors and thus protect bacteria from serum killing. Structural analyses show that PG0352 contains a N-terminal carbohydrate-binding module (CBM) and a C-terminal sialidase domain that exhibits a canonical six-bladed β-propeller sialidase fold with each blade composed of 3-4 antiparallel β-strands. Follow-up functional studies show that PG0352 forms monomers and is active in a broad range of pH. While PG0352 can remove both N-acetylneuraminic acid (Neu5Ac) and N-glycolyl-neuraminic acid (Neu5Gc), it has a higher affinity to Neu5Ac, the most abundant sialic acid in humans. Structural and functional analyses further demonstrate that the CBM binds to carbohydrates and serum glycoproteins. The results shown in this report provide new insights into understanding the role of sialidases in bacterial virulence and open a new avenue to investigate the molecular mechanisms of bacterial complement resistance.
- Department of Structural Biology, Jacobs School of Medicine and Biomedical Sciences, University of Buffalo, the State University of New York, Buffalo, New York, United States of America.
Organizational Affiliation: