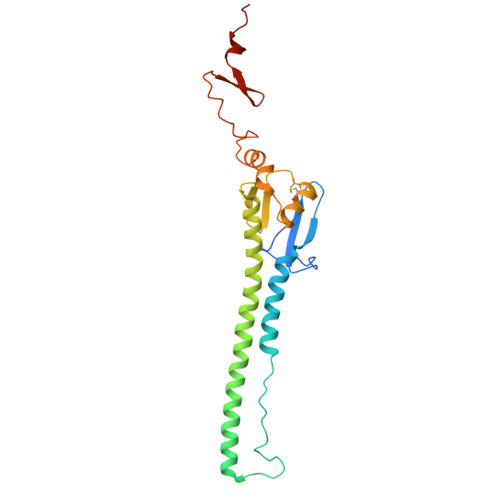
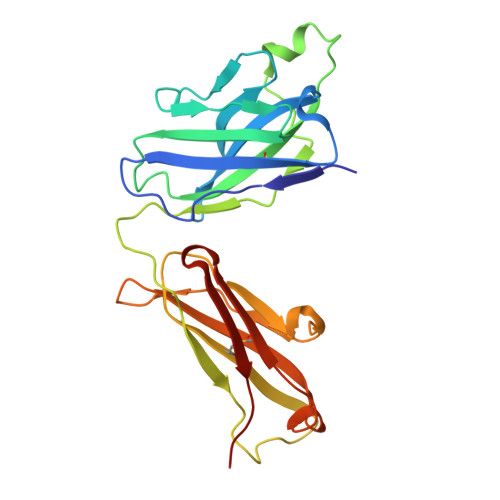
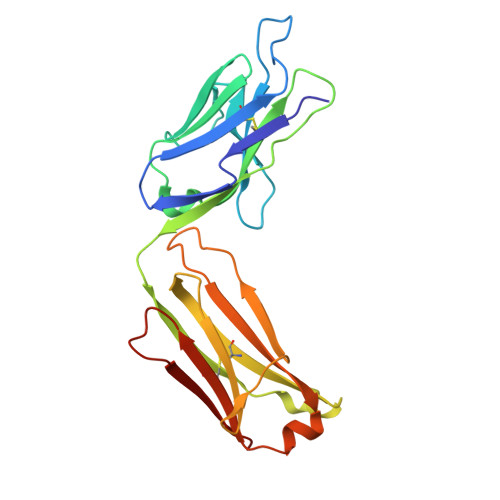
Distinct binding modes drive the broad neutralization profile of two persistent influenza hemagglutinin stem-specific antibody lineages.
Mantus, G.E., Cerutti, G., Chambers, M., Gillespie, R.A., Shimberg, G.D., Spangler, A., Gorman, J., Zhou, T., Shen, C.H., Kanekiyo, M., Kwong, P.D., Shapiro, L., Andrews, S.F.(2025) Structure 33: 869
- PubMed: 40112805
- DOI: https://doi.org/10.1016/j.str.2025.02.010
- Primary Citation of Related Structures:
8T1G, 8VEB, 8VED, 8VEE, 8VEF - PubMed Abstract:
Elicitation of antibodies to the influenza hemagglutinin stem is a critical part of universal influenza vaccine strategies. While numerous broadly reactive stem antibodies have been isolated, our understanding of how these antibodies mature within the human B cell repertoire is limited. Here, we isolated and tracked two stem-specific antibody lineages over a decade in a single participant that received multiple seasonal and pandemic influenza vaccinations. Despite similar binding and neutralization profiles, antibodies from these lineages utilized fundamentally different interactions to engage the central epitope on the influenza stem. Structural analysis of an unmutated common ancestor from one lineage identified critical residues that were the main drivers of increased affinity and breadth to group 1 influenza subtypes. These observations demonstrate the heterogeneous pathways by which stem-specific antibodies can mature within the human B cell repertoire.
- Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA.
Organizational Affiliation: