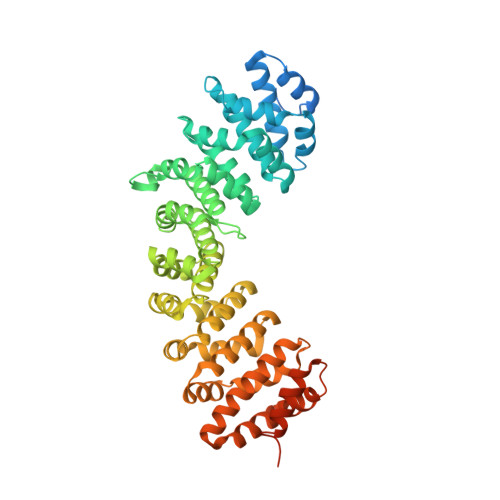
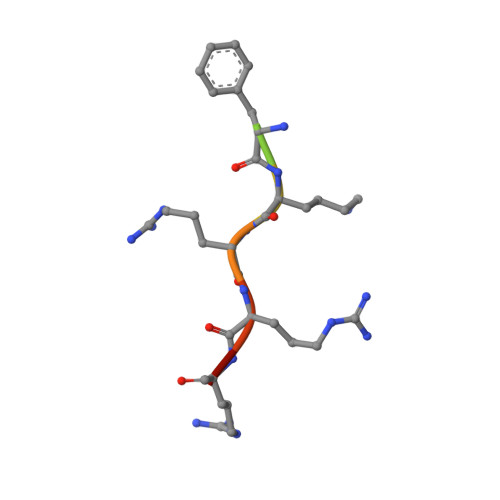
Structural and functional characterization of siadenovirus core protein VII nuclear localization demonstrates the existence of multiple nuclear transport pathways.
Athukorala, A., Donnelly, C.M., Pavan, S., Nematollahzadeh, S., Djossou, V.A., Nath, B., Helbig, K.J., Di Iorio, E., McSharry, B.P., Alvisi, G., Forwood, J.K., Sarker, S.(2024) J Gen Virol 105
- PubMed: 38261399
- DOI: https://doi.org/10.1099/jgv.0.001928
- Primary Citation of Related Structures:
8SV0 - PubMed Abstract:
Adenovirus protein VII (pVII) plays a crucial role in the nuclear localization of genomic DNA following viral infection and contains nuclear localization signal (NLS) sequences for the importin (IMP)-mediated nuclear import pathway. However, functional analysis of pVII in adenoviruses to date has failed to fully determine the underlying mechanisms responsible for nuclear import of pVII. Therefore, in the present study, we extended our analysis by examining the nuclear trafficking of adenovirus pVII from a non-human species, psittacine siadenovirus F (PsSiAdV). We identified a putative classical (c)NLS at pVII residues 120-128 ( 120 PGGFKRRRL 128 ). Fluorescence polarization and electrophoretic mobility shift assays demonstrated direct, high-affinity interaction with both IMPα2 and IMPα3 but not IMPβ. Structural analysis of the pVII-NLS/IMPα2 complex confirmed a classical interaction, with the major binding site of IMPα occupied by K 124 of pVII-NLS. Quantitative confocal laser scanning microscopy showed that PsSiAdV pVII-NLS can confer IMPα/β-dependent nuclear localization to GFP. PsSiAdV pVII also localized in the nucleus when expressed in the absence of other viral proteins. Importantly, in contrast to what has been reported for HAdV pVII, PsSiAdV pVII does not localize to the nucleolus. In addition, our study demonstrated that inhibition of the IMPα/β nuclear import pathway did not prevent PsSiAdV pVII nuclear targeting, indicating the existence of alternative pathways for nuclear localization, similar to what has been previously shown for human adenovirus pVII. Further examination of other potential NLS signals, characterization of alternative nuclear import pathways, and investigation of pVII nuclear targeting across different adenovirus species is recommended to fully elucidate the role of varying nuclear import pathways in the nuclear localization of pVII.
- Department of Microbiology, Anatomy, Physiology, and Pharmacology, School of Agriculture, Biomedicine and Environment, La Trobe University, Melbourne, VIC 3086, Australia.
Organizational Affiliation: