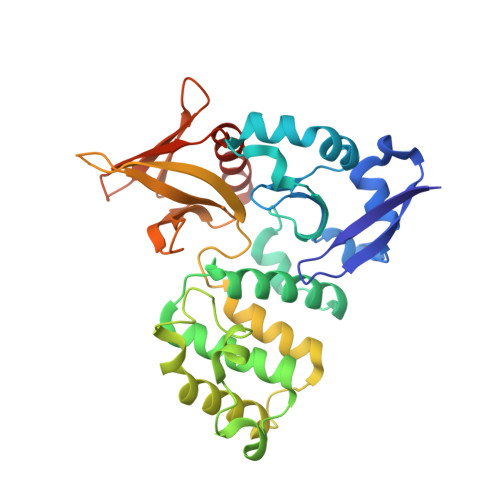
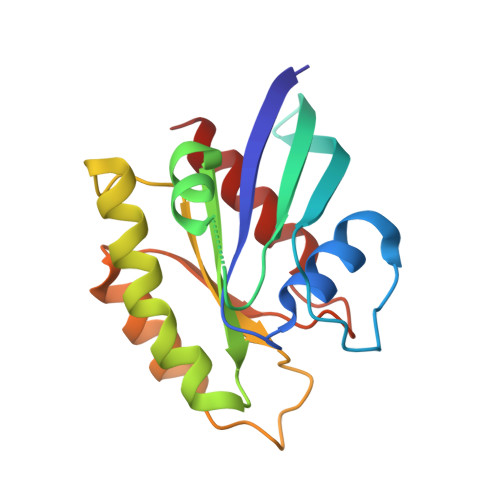
Targeted Reversible Covalent Modification of a Noncatalytic Lysine of the Krev Interaction Trapped 1 Protein Enables Site-Directed Screening for Protein-Protein Interaction Inhibitors.
Francisco, K.R., Bruystens, J., Varricchio, C., McCurdy, S., Wu, J., Lopez-Ramirez, M.A., Ginsberg, M., Caffrey, C.R., Brancale, A., Gingras, A.R., Hixon, M.S., Ballatore, C.(2023) Acs Pharmacol Transl Sci 6: 1651-1658
- PubMed: 37974623
- DOI: https://doi.org/10.1021/acsptsci.3c00156
- Primary Citation of Related Structures:
8SU8, 8T09, 8T7V - PubMed Abstract:
The covalent reversible modification of proteins is a validated strategy for the development of probes and candidate therapeutics. However, the covalent reversible targeting of noncatalytic lysines is particularly challenging. Herein, we characterize the 2-hydroxy-1-naphthaldehyde (HNA) fragment as a targeted covalent reversible ligand of a noncatalytic lysine (Lys 720 ) of the Krev interaction trapped 1 (KRIT1) protein. We show that the interaction of HNA with KRIT1 is highly specific, results in prolonged residence time of >8 h, and inhibits the Heart of glass 1 (HEG1)-KRIT1 protein-protein interaction (PPI). Screening of HNA derivatives identified analogs exhibiting similar binding modes as the parent fragment but faster target engagement and stronger inhibition activity. These results demonstrate that HNA is an efficient site-directing fragment with promise in developing HEG1-KRIT1 PPI inhibitors. Further, the aldimine chemistry, when coupled with templating effects that promote proximity, can produce a long-lasting reversible covalent modification of noncatalytic lysines.
- Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman Drive, La Jolla, California 92093, United States.
Organizational Affiliation: