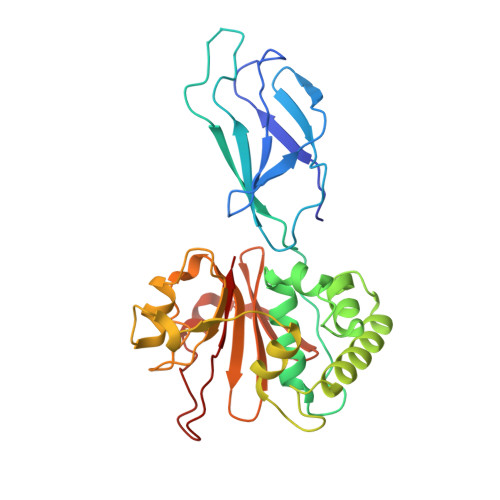
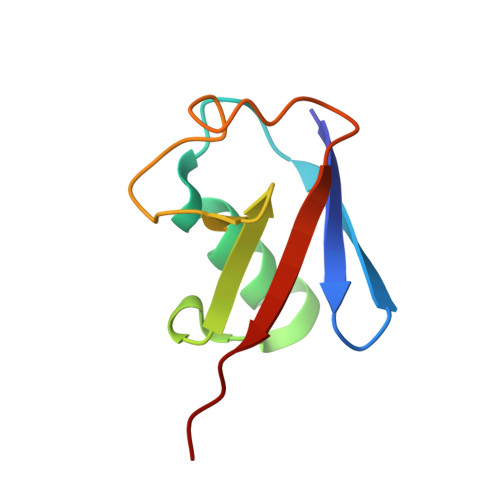
Bacterial esterases reverse lipopolysaccharide ubiquitylation to block host immunity.
Szczesna, M., Huang, Y., Lacoursiere, R.E., Bonini, F., Pol, V., Koc, F., Ward, B., Geurink, P.P., Pruneda, J.N., Thurston, T.L.M.(2024) Cell Host Microbe 32: 913-924.e7
- PubMed: 38870903
- DOI: https://doi.org/10.1016/j.chom.2024.04.012
- Primary Citation of Related Structures:
8SSI - PubMed Abstract:
Aspects of how Burkholderia escape the host's intrinsic immune response to replicate in the cell cytosol remain enigmatic. Here, we show that Burkholderia has evolved two mechanisms to block the activity of Ring finger protein 213 (RNF213)-mediated non-canonical ubiquitylation of bacterial lipopolysaccharide (LPS), thereby preventing the initiation of antibacterial autophagy. First, Burkholderia's polysaccharide capsule blocks RNF213 association with bacteria and second, the Burkholderia deubiquitylase (DUB), TssM, directly reverses the activity of RNF213 through a previously unrecognized esterase activity. Structural analysis provides insight into the molecular basis of TssM esterase activity, allowing it to be uncoupled from its isopeptidase function. Furthermore, a putative TssM homolog also displays esterase activity and removes ubiquitin from LPS, establishing this as a virulence mechanism. Of note, we also find that additional immune-evasion mechanisms exist, revealing that overcoming this arm of the host's immune response is critical to the pathogen.
- Department of Infectious Disease, Centre for Bacterial Resistance Biology, Imperial College London, London SW7 2AZ, UK.
Organizational Affiliation: