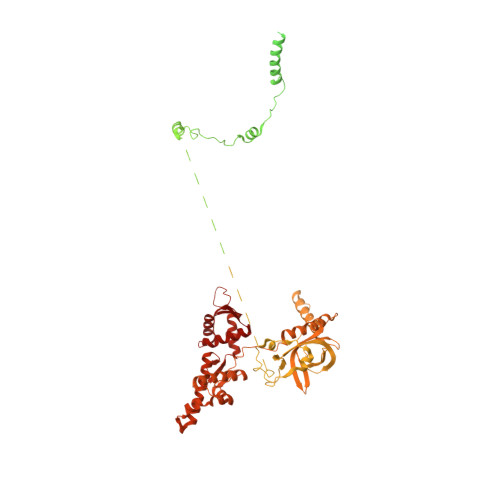
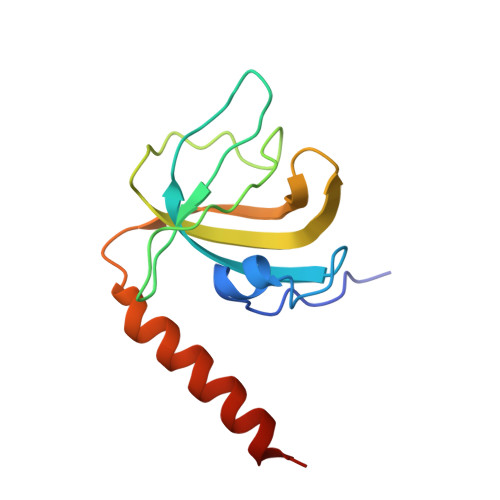
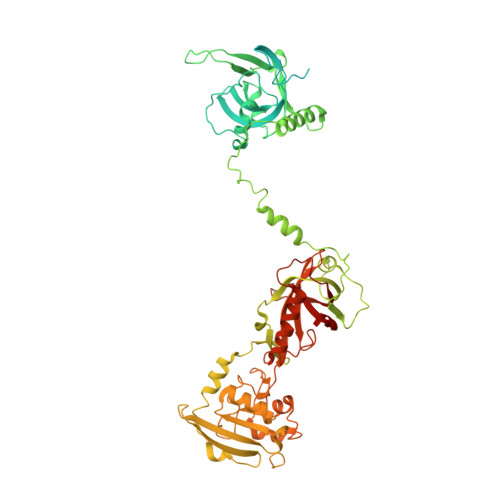
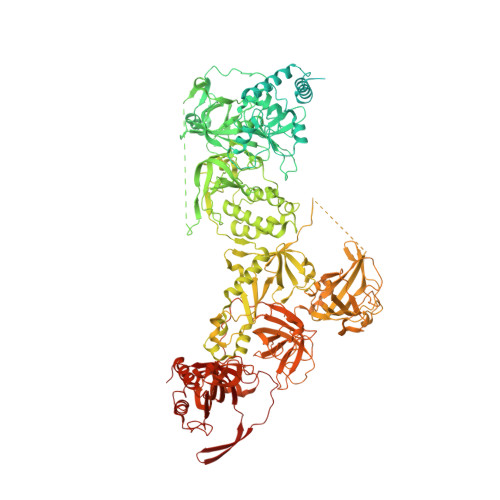POT1 recruits and regulates CST-Pol alpha /primase at human telomeres.
Cai, S.W., Takai, H., Zaug, A.J., Dilgen, T.C., Cech, T.R., Walz, T., de Lange, T.(2024) Cell 187: 3638-3651.e18
- PubMed: 38838667
- DOI: https://doi.org/10.1016/j.cell.2024.05.002
- Primary Citation of Related Structures:
8SOJ, 8SOK - PubMed Abstract:
Telomere maintenance requires the extension of the G-rich telomeric repeat strand by telomerase and the fill-in synthesis of the C-rich strand by Polα/primase. At telomeres, Polα/primase is bound to Ctc1/Stn1/Ten1 (CST), a single-stranded DNA-binding complex. Like mutations in telomerase, mutations affecting CST-Polα/primase result in pathological telomere shortening and cause a telomere biology disorder, Coats plus (CP). We determined cryogenic electron microscopy structures of human CST bound to the shelterin heterodimer POT1/TPP1 that reveal how CST is recruited to telomeres by POT1. Our findings suggest that POT1 hinge phosphorylation is required for CST recruitment, and the complex is formed through conserved interactions involving several residues mutated in CP. Our structural and biochemical data suggest that phosphorylated POT1 holds CST-Polα/primase in an inactive, autoinhibited state until telomerase has extended the telomere ends. We propose that dephosphorylation of POT1 releases CST-Polα/primase into an active state that completes telomere replication through fill-in synthesis.
- Laboratory of Cell Biology and Genetics, The Rockefeller University, New York, NY 10065, USA; Laboratory of Molecular Electron Microscopy, The Rockefeller University, New York, NY 10065, USA.
Organizational Affiliation: