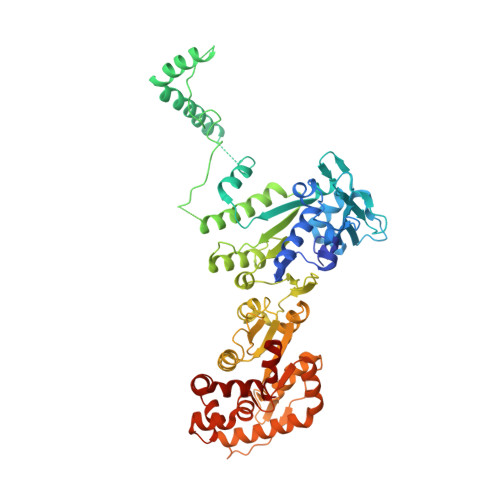
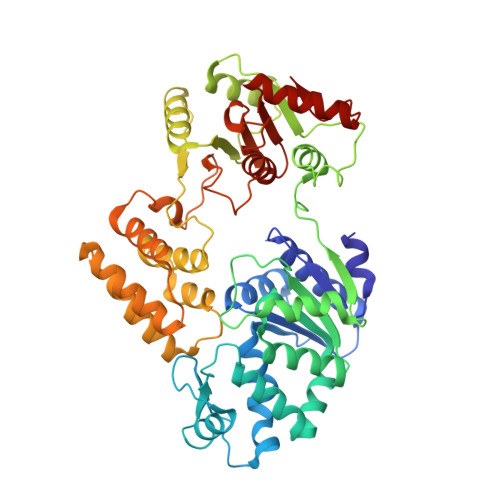
Structural basis of Gabija anti-phage defence and viral immune evasion.
Antine, S.P., Johnson, A.G., Mooney, S.E., Leavitt, A., Mayer, M.L., Yirmiya, E., Amitai, G., Sorek, R., Kranzusch, P.J.(2024) Nature 625: 360-365
- PubMed: 37992757
- DOI: https://doi.org/10.1038/s41586-023-06855-2
- Primary Citation of Related Structures:
8SM3, 8U7I - PubMed Abstract:
Bacteria encode hundreds of diverse defence systems that protect them from viral infection and inhibit phage propagation 1-5 . Gabija is one of the most prevalent anti-phage defence systems, occurring in more than 15% of all sequenced bacterial and archaeal genomes 1,6,7 , but the molecular basis of how Gabija defends cells from viral infection remains poorly understood. Here we use X-ray crystallography and cryo-electron microscopy (cryo-EM) to define how Gabija proteins assemble into a supramolecular complex of around 500 kDa that degrades phage DNA. Gabija protein A (GajA) is a DNA endonuclease that tetramerizes to form the core of the anti-phage defence complex. Two sets of Gabija protein B (GajB) dimers dock at opposite sides of the complex and create a 4:4 GajA-GajB assembly (hereafter, GajAB) that is essential for phage resistance in vivo. We show that a phage-encoded protein, Gabija anti-defence 1 (Gad1), directly binds to the Gabija GajAB complex and inactivates defence. A cryo-EM structure of the virally inhibited state shows that Gad1 forms an octameric web that encases the GajAB complex and inhibits DNA recognition and cleavage. Our results reveal the structural basis of assembly of the Gabija anti-phage defence complex and define a unique mechanism of viral immune evasion.
- Department of Microbiology, Harvard Medical School, Boston, MA, USA.
Organizational Affiliation: