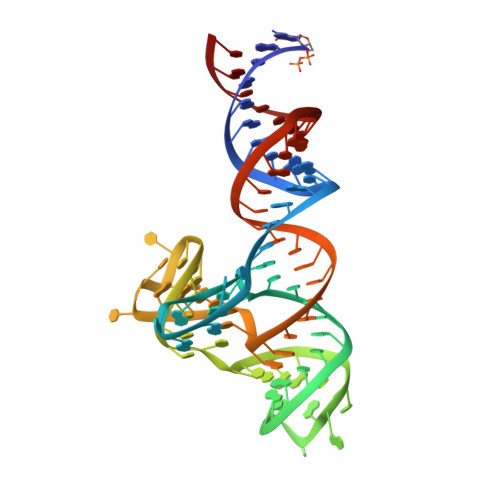
Crystal structure of a cap-independent translation enhancer RNA.
Lewicka, A., Roman, C., Jones, S., Disare, M., Rice, P.A., Piccirilli, J.A.(2023) Nucleic Acids Res 51: 8891-8907
- PubMed: 37548413
- DOI: https://doi.org/10.1093/nar/gkad649
- Primary Citation of Related Structures:
8SH5 - PubMed Abstract:
In eukaryotic messenger RNAs, the 5' cap structure binds to the translation initiation factor 4E to facilitate early stages of translation. Although many plant viruses lack the 5' cap structure, some contain cap-independent translation elements (CITEs) in their 3' untranslated region. The PTE (Panicum mosaic virus translation element) class of CITEs contains a G-rich asymmetric bulge and a C-rich helical junction that were proposed to interact via formation of a pseudoknot. SHAPE analysis of PTE homologs reveals a highly reactive guanosine residue within the G-rich region proposed to mediate eukaryotic initiation factor 4E (eIF4E) recognition. Here we have obtained the crystal structure of the PTE from Pea enation mosaic virus 2 (PEMV2) RNA in complex with our structural chaperone, Fab BL3-6. The structure reveals that the G-rich and C-rich regions interact through a complex network of interactions distinct from those expected for a pseudoknot. The motif, which contains a short parallel duplex, provides a structural mechanism for how the guanosine is extruded from the core stack to enable eIF4E recognition. Homologous PTE elements harbor a G-rich bulge and a three-way junction and exhibit covariation at crucial positions, suggesting that the PEMV2 tertiary architecture is conserved among these homologs.
- Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL 60637, USA.
Organizational Affiliation: