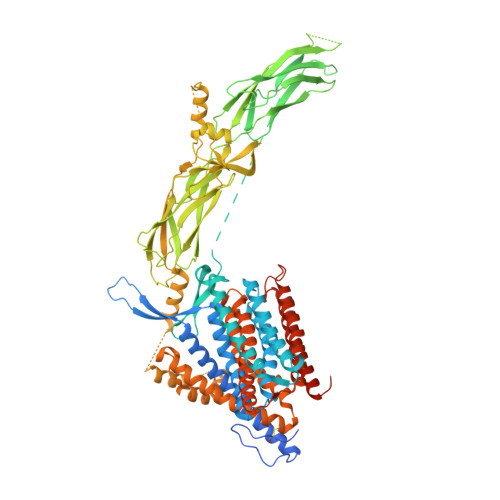
Structural mechanisms of PIP 2 activation and SEA0400 inhibition in human cardiac sodium-calcium exchanger NCX1.
Xue, J., Zeng, W., John, S., Attiq, N., Ottolia, M., Jiang, Y.(2025) Elife 14
- PubMed: 40433952
- DOI: https://doi.org/10.7554/eLife.105396
- Primary Citation of Related Structures:
8SGI, 9IV8 - PubMed Abstract:
Na + /Ca 2+ exchangers (NCXs) transport Ca 2+ across the plasma membrane in exchange for Na + and play a vital role in maintaining cellular Ca 2+ homeostasis. Our previous structural study of human cardiac NCX1 (HsNCX1) reveals the overall architecture of the eukaryotic exchanger and the formation of the inactivation assembly by the intracellular regulatory domain that underlies the cytosolic Na + -dependent inactivation and Ca 2+ activation of NCX1. Here, we present the cryo-EM structures of HsNCX1 in complex with a physiological activator phosphatidylinositol 4,5-bisphosphate (PIP 2 ), or pharmacological inhibitor SEA0400, that enhances the inactivation of the exchanger. We demonstrate that PIP 2 binding stimulates NCX1 activity by inducing a conformational change at the interface between the transmembrane (TM) and cytosolic domains that destabilizes the inactivation assembly. In contrast, SEA0400 binding in the TM domain of NCX1 stabilizes the exchanger in an inward-facing conformation that facilitates the formation of the inactivation assembly, thereby promoting the Na + -dependent inactivation of NCX1. Thus, this study reveals the structural basis of PIP 2 activation and SEA0400 inhibition of NCX1 and provides some mechanistic understandings of cellular regulation and pharmacology of NCX family proteins.
- Department of Physiology, The University of Texas Southwestern Medical Center, Dallas, United States.
Organizational Affiliation: