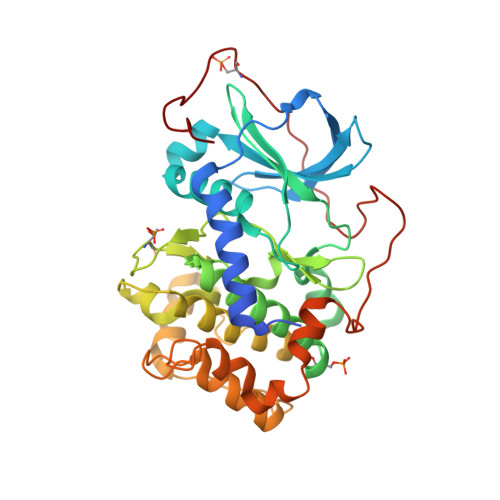
Discovery and Characterization of Selective, First-in-Class Inhibitors of Citron Kinase.
Maw, J.J., Coker, J.A., Arya, T., Goins, C.M., Sonawane, D., Han, S.H., Rees, M.G., Ronan, M.M., Roth, J.A., Wang, N.S., Heemers, H.V., Macdonald, J.D., Stauffer, S.R.(2024) J Med Chem 67: 2631-2666
- PubMed: 38330278
- DOI: https://doi.org/10.1021/acs.jmedchem.3c01807
- Primary Citation of Related Structures:
8SF8 - PubMed Abstract:
Citron kinase (CITK) is an AGC-family serine/threonine kinase that regulates cytokinesis. Despite knockdown experiments implicating CITK as an anticancer target, no selective CITK inhibitors exist. We transformed a previously reported kinase inhibitor with weak off-target CITK activity into a first-in-class CITK chemical probe, C3TD879 . C3TD879 is a Type I kinase inhibitor which potently inhibits CITK catalytic activity (biochemical IC 50 = 12 nM), binds directly to full-length human CITK in cells (NanoBRET K d < 10 nM), and demonstrates favorable DMPK properties for in vivo evaluation. We engineered exquisite selectivity for CITK (>17-fold versus 373 other human kinases), making C3TD879 the first chemical probe suitable for interrogating the complex biology of CITK. Our small-molecule CITK inhibitors could not phenocopy the effects of CITK knockdown in cell proliferation, cell cycle progression, or cytokinesis assays, providing preliminary evidence that the structural roles of CITK may be more important than its kinase activity.
- Center for Therapeutics Discovery, Lerner Research Institute, Cleveland Clinic, Cleveland, Ohio 44195, United States.
Organizational Affiliation: